31 October 2022: Clinical Research
hOGG1 rs1052133 Polymorphism and Prostate Cancer Risk: A Chinese Case-Control Study and Meta-Analysis
Hanjiang Xu123BCDEF*, Meng Zhang123BCDEF, Zongyao Hao123AC, Chaozhao Liang123ADOI: 10.12659/MSM.938012
Med Sci Monit 2022; 28:e938012
Abstract
BACKGROUND: We performed a case-control study and an updated meta-analysis to assess the relationship between the hOGG1 rs1052133 polymorphism and prostate cancer (PCa) risk.
MATERIAL AND METHODS: We recruited 160 PCa cases and 243 healthy controls. For the meta-analysis, relevant studies were recruited from diverse databases up to April 2022. Genetic risk was evaluated by using an odds ratio (OR) with a corresponding 95% confidence interval (95% CI). The genotypes of this polymorphism were genotyped via the SNaPshot genotyping method.
RESULTS: In the case-control study, we failed to identify any association between the hOGG1 rs1052133 polymorphism and PCa risk. Negative results were also obtained when stratified analyses were performed based on the patient’s prostatic-specific antigen (PSA) level and Gleason score, as well as tumor, node, and metastasis (TNM) stage. To enlarge the sample size, we performed a restricted updated meta-analysis by recruiting 10 case-control studies (including the current one), and the results suggested that genotypes of rs1052133 polymorphism were significantly associated with an elevated risk of PCa in 2 genetic models – the heterozygote and dominant models. In the stratification analysis by population ethnicity, a significant association of this polymorphism with susceptibility to PCa was found both in the Asian populations and White populations.
CONCLUSIONS: Our case-control and updated meta-analysis study suggest that the hOGG1 rs1052133 polymorphism is a susceptibility factor for PCa, but still needs to be further verified in the Chinese population.
Keywords: Oxoguanine Glycosylase 1, Human, Polymorphism, Genetic, Prostatic Neoplasms, Humans, Male, Case-Control Studies, China, DNA Glycosylases, Genetic Predisposition to Disease, Genotype, Odds Ratio, Polymorphism, Single Nucleotide, Risk Factors
Background
The role of DNA damage in tumor formation and development is well established [1]. As a result of oxidative stress, inflammation, or environmental carcinogens, DNA damage can accumulate in the prostate, possibly increasing the risk for prostate cancer (PCa) [2–4]. The base excision repair (BER) pathway is the most often-used approach for removing minor damage from DNA. Despite its structural independence, it is also crucial in cellular defense against many types of DNA lesions [5,6].
The human 8-oxoguanine glycosylase 1 (hOGG1) gene, located on chromosome 3p26, plays an essential role in DNA damage repair by initiating the BER pathway [7–9]. Rs1052133 is a commonly occurring polymorphism in hOGG1, and also referred to as Ser326Cys polymorphism. Cysteine can be substituted by serine at 326 amino acids of hOGG1 protein. Notably, there is an altered susceptibility for diverse cancer types related to its genotypes [10–12].
In recent years, the hOGG1 rs1052133 polymorphism has been often investigated in relation to PCa across diverse ethnic populations, but the results were inconsistent [13–24]. Here, we assessed whether genotypes of the hOGG1 rs1052133 polymorphism were related to an elevated PCa risk in a Chinese Han population. Additionally, we performed a comprehensive updated meta-analysis of published investigations to determine the exact relationship between them.
Material and Methods
Case-Control Study
SELECTION OF ELIGIBLE PCA CASES AND HEALTHY CONTROLS:
We enrolled 160 PCa patients and 243 healthy controls from the First Affiliated Hospital of Anhui Medical University between January 2016 and December 2020. A biopsy or postoperative pathology of transurethral resection prostate (TURP) was used to diagnose PCa patients within 1 year of enrollment. In the present study, patients pathologically confirmed to have prostate adenocarcinoma were enrolled for subsequent analysis. Prior to invasive manipulation, PSA levels were assessed, excluding those patients who had received endocrine therapy. Gleason grades were determined by biopsy or radical prostatectomy. Based on the radical prostatectomy specimens or the results of computed tomography, magnetic resonance imaging, or bone scans, the tumor stage was determined. All the controls were cancer-free individuals who underwent regular physical examinations at the hospital. Study controls with serum PSA levels >4 ng/ml were excluded to rule out prostate cancer. The investigators excluded patients with incomplete medical records. The Research Ethics Committee at the First Affiliated Hospital of Anhui Medical University approved the study. Furthermore, each participant signed an informed consent form in duplicate.
GENOTYPING: Commercially available DNA extraction kits (Cat. No. 51106; Qiagen, Inc., Valencia, CA) were used to extract DNA from each participant’s blood sample. The site sequence of the hOGG1 rs1052133 polymorphism was obtained from the NCBI db SNP database. For each DNA sample, the hOGG1 rs1052133 polymorphism was detected by using the SNaPshot SNP assay by Shanghai Tianhao company (Shanghai Tianhao Industrial Co., Ltd., Shanghai, China) [25,26]. We used the Multiplex SNaPshot technology (Applied Biosystems, Foster City, CA, USA) to determine the genotype of hOGG1 gene. Primer3 online software (v.0.4.0) (http://frodo.wi.mit.edu/primer3/) was used to design the primers for PCR and the SNaPshot extension reactions based on the sequences provided in dbSNP (http://www.ncbi.nlm.nih.gov/SNP) (rs1052133F: CCAGGTGGCCCTAAAGGACTCT; rs1052133R: GTGGGGATGGGGAGAGAGAAGT; primer extension: TGGCTCCTGAGCATGGCGG). The products were sequenced by ABI3130XL Sequencer (Applied Biosystems, Foster City, CA, USA), and GeneMapper 4.0 (Applied Biosystems, Foster City, CA, USA) was used to analyze the data. The type of nucleotide presented in the SNP locus was used to determine the genotype of each sample, which was analyzed by 1 or 2 distinct colored peaks on the graph.
STATISTICAL ANALYSIS FOR REAL-WORLD COHORT:
Differences between the 2 groups were determined using the 2-tailed unpaired
META-ANALYSIS:
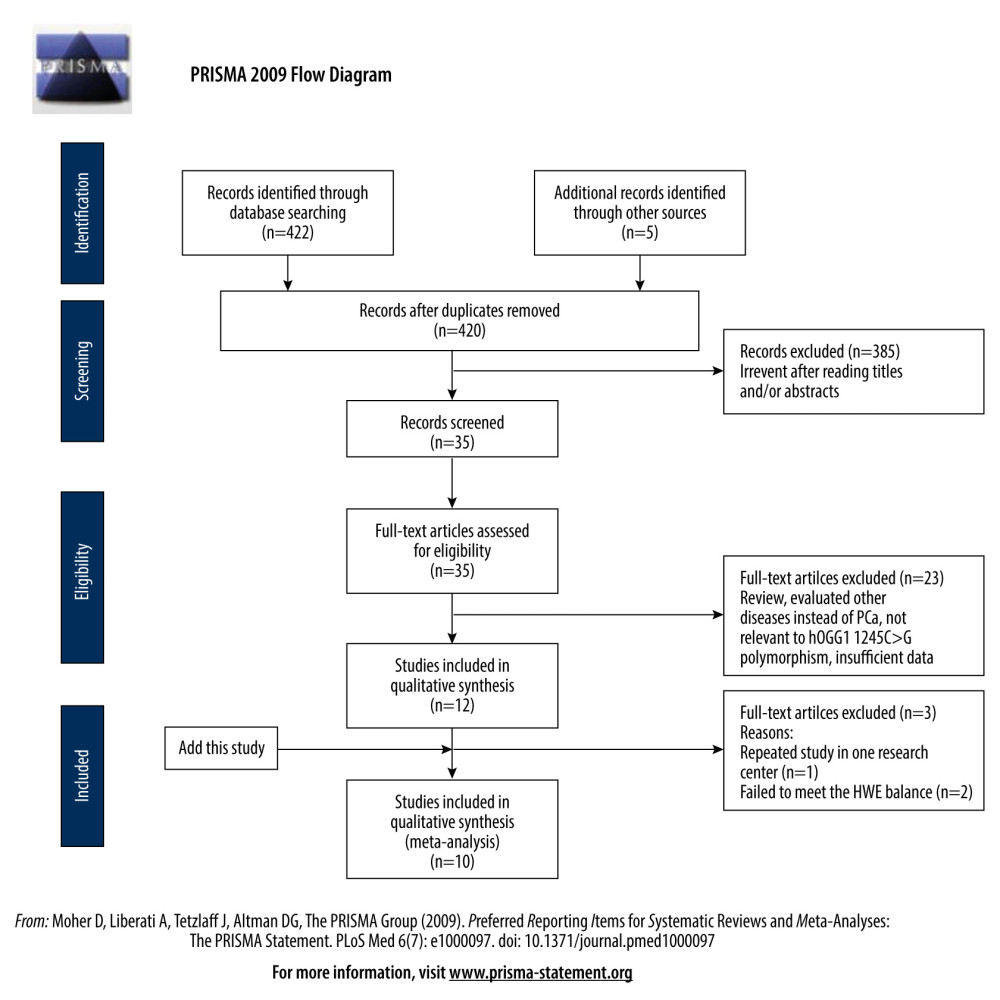
The meta-analysis was performed following the PRISMA Statement [27]. Figure 1 shows the whole process of the study.
PUBLICATION SEARCH:
Our electronic literature search utilized PubMed, Web of Science, and EMBASE to find all eligible studies that evaluated the risk associated between the hOGG1 rs1052133 polymorphism and the risk of PCa up to April 2022. In this search, the following MeSH terms and keywords were employed: (“PCa” OR “prostate cancer” OR “prostate tumor” OR “prostate adenocarcinoma” OR “prostate neoplasm”) AND (“hOGG1” OR “human 8-oxoguanine DNA glycosylase 1”) AND (“gene” OR “polymorphism” OR “variant” OR “allele” OR “mutation”) AND (“rs1052133” OR “Ser326Cys” OR “1245C>G” OR “C8069G”). The entire search process was conducted in English. If the same population was presented in multiple publications, only the publication with the largest sample size was enrolled.
INCLUSION AND EXCLUSION CRITERIA:
Eligible studies were recruited to the meta-analysis when the following criteria were reached: (1) a case-control study; (2) a cohort study; (3) evaluation of the presence of the rs1052133 polymorphism and the risk of PCa; (3) adequate data were available; and (4) genotype distributions following Hardy-Weinberg equilibrium (HWE).
Accordingly, we excluded studies with the following characteristics: (1) cross-sectional cohort studies published in other languages, non-original studies, dissertations, and thesis studies; (2) insufficient data or studies lacking genotype distribution data; and (3) study was not relevant to hOGG1 rs1052133 or the PCa polymorphism. Author(s), publication date, country, ethnicity, examined genes (SNPs), sample number, genotyping method used, genotypic frequencies, and allelic frequencies were collected. Two reviewers (HJX and MZ) independently evaluated the qualities of enrolled studies in our meta-analysis. The qualities of the included studies were assessed by the Newcastle-Ottawa Scale (NOS) score method [28] from the different aspects, including selection, comparability, exposure/outcome (Supplementary Table 1).
META-ANALYSIS:
Genotype models (the heterozygote, homozygote, dominant, and recessive genetic models) were employed to determine the correlation of the hOGG1 rs1052133 polymorphism with PCa risk by using the crude OR with related 95% CI.
We determined the methods to calculate the OR from the χ2-based Q-test and checked the heterogeneity of the current study. If
Results
Case-Control Study
DEMOGRAPHIC FEATURES: The average age in the PCa group was 70.09±7.14 years compared to 71.30±8.80 years in the negative control group (t test, P=0.148). In the PCa group, 46 (28.75%) subjects showed a PSA level <10 ng/ml, 50 (31.25%) subjects showed a PSA level of 10–20 ng/ml, and 64 (40.00%) subjects showed a PSA level >20 ng/ml. The number of PCa cases whose Gleason score ≤7 was 99 (61.88%), and the number with a Gleason score >7 was 61 (38.12%). Moreover, there were 119 (74.38%) patients and 41 (25.62%) patients with stage <T3 and ≥T3, respectively. We summarized these features in Table 1.
ASSOCIATION OF HOGG1 GENOTYPES WITH PCA RISK: The prevalence of the hOGG1 rs1052133 polymorphism in healthy controls followed HWE (P=0.380). As indicated, hOGG1 polymorphism was not correlated with an increased risk of PCa in the 4 genetic models in our case-control study (Table 2). We further analyzed the correlation between subgroups (PSA level, Gleason score, and TNM stage) in PCa cases to validate the correlation with the predictive risk of PCa (Table 3). However, none of these differences were statistically significant, which was possibly due to the limited sample size.
ELIGIBLE STUDIES: Twelve eligible studies were found by using the selected keywords (Table 4) [13–24]; however, 1 repeated study was removed [19], and 2 studies that failed to meet the HWE balance were excluded [20,21]. Finally, 10 studies were included in the meta-analysis, including the data that were generated from the present case-control study.
META-ANALYSIS: The detailed associations of the hOGG1 rs1052133 polymorphism with PCa susceptibility in all of the genetic models are presented in Table 5. The results showed that rs1052133 polymorphism was significantly related to an elevated PCa risk in 2 genetic models, including heterozygote (OR: 1.172, 95% CI: 1.033–1.330, P=0.014) and dominant models (OR: 1.227, 95% CI: 1.003–1.500, P=0.047). In the stratification analysis by ethnicity, we observed a significant association of the rs1052133 polymorphism with susceptibility to PCa risk in Asian population in 3 genetic models, including allele (OR: 1.217, 95% CI: 1.052–1.409, p=0.008), heterozygote (OR: 1.593, 95% CI: 1.160–2.187, P=0.004), and dominant models (OR: 1.299, 95% CI: 1.030–1.638, P=0.027), and in White populations in all of the genetic models, but it should be noted that there were 2 studies with small sample sizes that were included (Table 5).
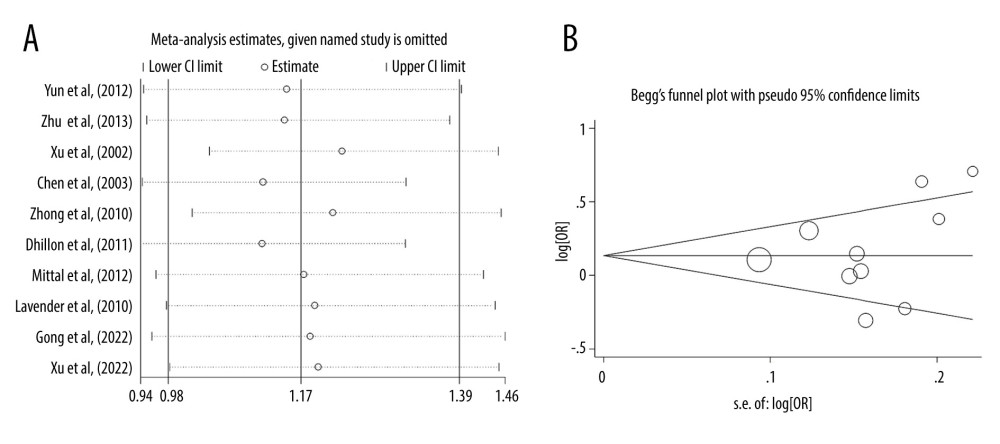
Moreover, the findings were stable and robust, which was supported by the sensitivity analysis after removing any of the individual studies (Figure 2A). Both the plot of Begg’s funnel (Figure 2B) and test of Egger’s regression analysis (P>|t|=0.478) suggested that no publication bias existed.
Discussion
In developed and developing countries alike, PCa is a malignancy that is strongly influenced by genetic factors. According to Yamane et al [29], the hOGG1 gene is an integral component of the DNA repair pathway. hOGG1-Cys326 (also termed rs1052133) was much less effective at preventing mutations in human cells than hOGG1-Ser326. Studies have suggested that the hOGG1 rs1052133 polymorphism may be important in determining PCa susceptibility. There is disagreement over the relationship between the rs1052133 polymorphism in hOGG1 and PCa susceptibility.
The first evidence for the increased prostate cancer risk of men with the CC genotype (Ser326) was presented by Xu et al [13] in 2002, which compared this genotype with homozygous GG men. Similar findings have been confirmed in the Korean population [22] and White population [14]. Moreover, Dhillon et al [17] found that only G allele of the rs1052133 polymorphism was related to an elevated risk of PCa. In 2 studies that were conducted in the Chinese Han population, one study demonstrated that the hOGG1 rs1052133 polymorphism was correlated with an enhanced malignant potential of PCa [23], whereas the other study reported that this polymorphism was more associated with the risk of low-grade prostate cancer [24]. Lavender et al. [16] and Mittal et al. [18] proposed contrasting opinions that they failed to observe any significant association between the HOGG1 genotypes and PCa risk. These discordant and conflicting results may be due to limited sample sizes and different genetic backgrounds.
Several meta-analysis studies have made efforts to test the relationship of the hOGG1 rs1052133 polymorphism with PCa risk. Notably, our results are slightly different from those published studies via the improvement of some flaws [30–32]. In 2012, Zhu et al [30] performed a meta-analysis study, and they found that the rs1052133 polymorphism was related to an elevated risk of PCa in both Whites and Asians. However, in the study by Agalliu et al. [33], they focused on the rs3218997 polymorphism and PCa risk, which was incorrectly recruited and synthesized in the study by Zhu et al. [30]. In 2015, Chen et al [32] performed an updated meta-analysis comprising 11 studies. However, in the pooled analysis, their results failed to show any correlation between the rs1052133 polymorphism and PCa risk. Significant associations were only found in the Asian population. Two studies with duplicated samples were both enrolled, which would cause a potential bias [18,19]. Furthermore, for those studies that failed, the HWE balance should be removed from the pooled analysis. Thus, we aimed to perform a case-control study and comprehensive updated meta-analysis to reveal the associations. In our case-control study, we failed to identify any positive results between the genotypes and PCa risk, which is a result consistent with most previous studies. Notably, based on the newly generated data from our case-control study, we performed a more rigorous and updated meta-analysis to determine the association of the rs1052133 polymorphism and PCa risk. Finally, we recruited 10 case-control studies, including 2218 PCa cases and 2946 controls. As a result, we obtained a positive correlation between the hOGG1 rs1052133 polymorphism and an elevated risk of PCa. Our meta-analysis identified a statistically significant correlation between the rs1052133 and PCa risk.
However, there were several shortcomings to be addressed in this study. First, this was a case-control study based in a hospital; thus, a selection bias should be considered. Additionally, our case-control study only analyzed the Chinese Han population with small sample size; therefore, more research is needed to confirm these findings. Finally, the biological function of this polymorphism during the progression of PCa has not been investigated.
Conclusions
Our study indicates that the rs1052133 polymorphism in hOGG1 is related to an elevated risk of PCa. However, more studies are necessary to examine their precise effects and the genuine associations between them in different countries and ethnicities.
Tables
Table 1. Comparison of clinical pathological characteristics between prostate cancer cases and controls.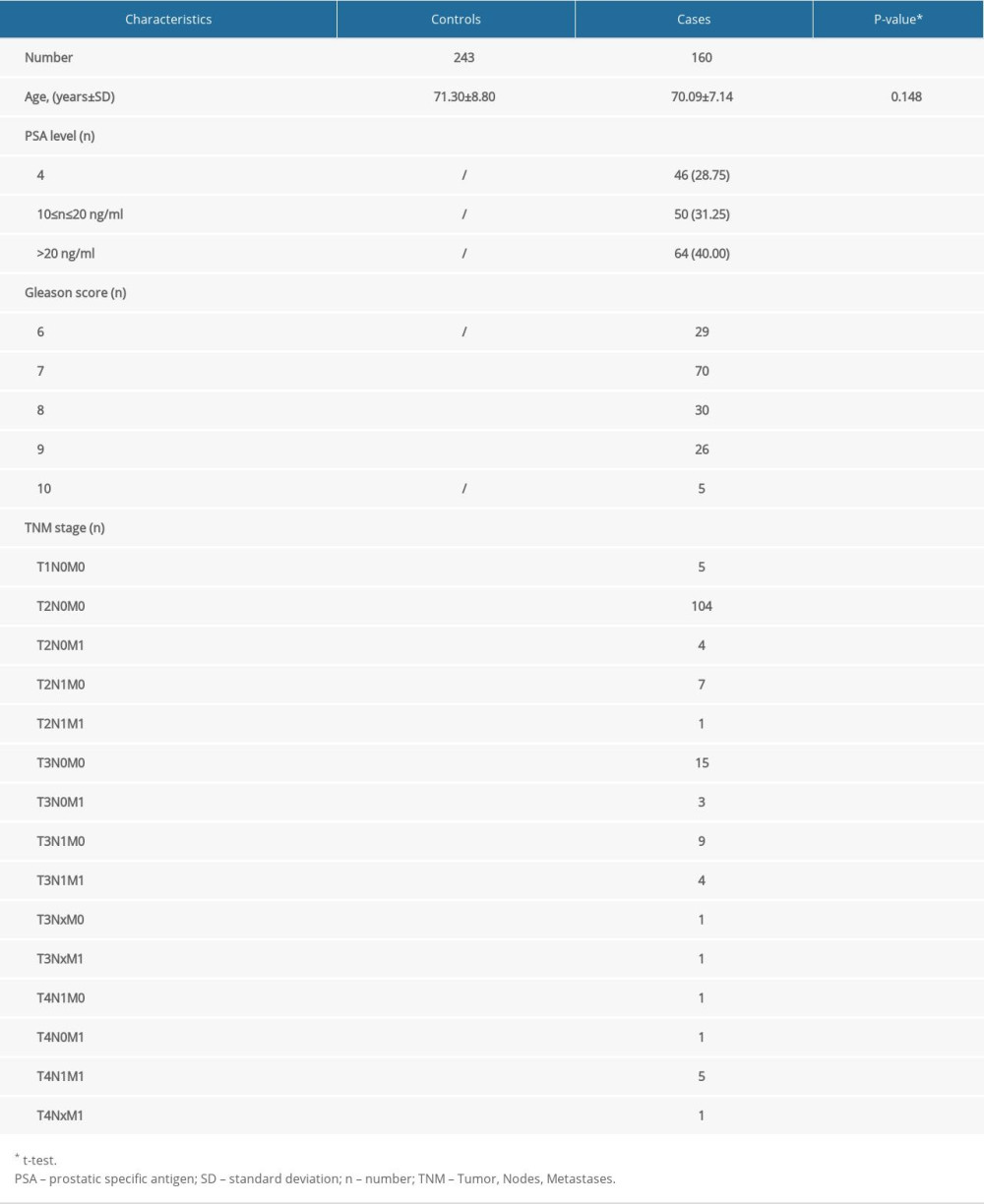 Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls.
Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls.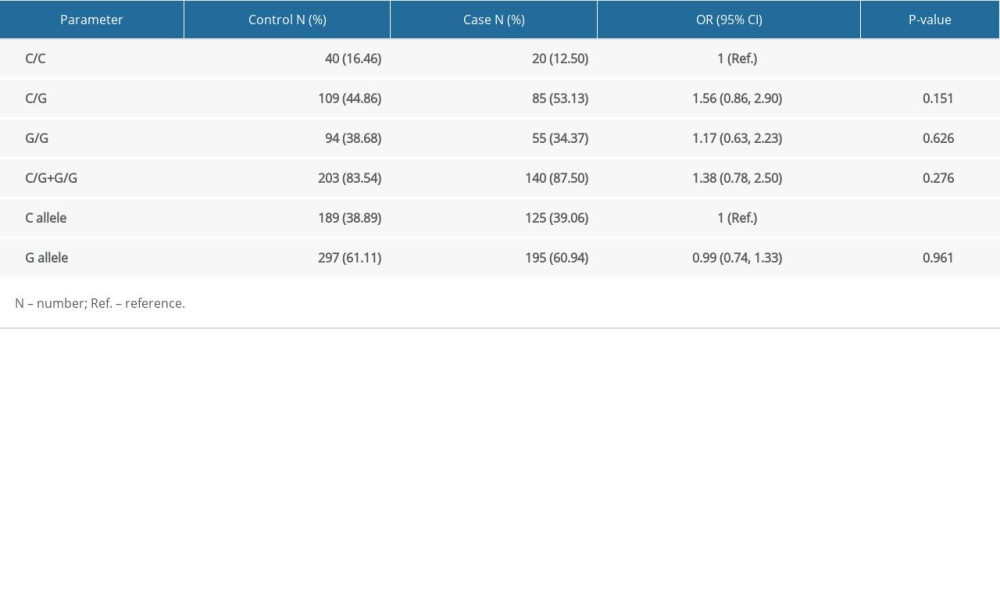 Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer.
Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer.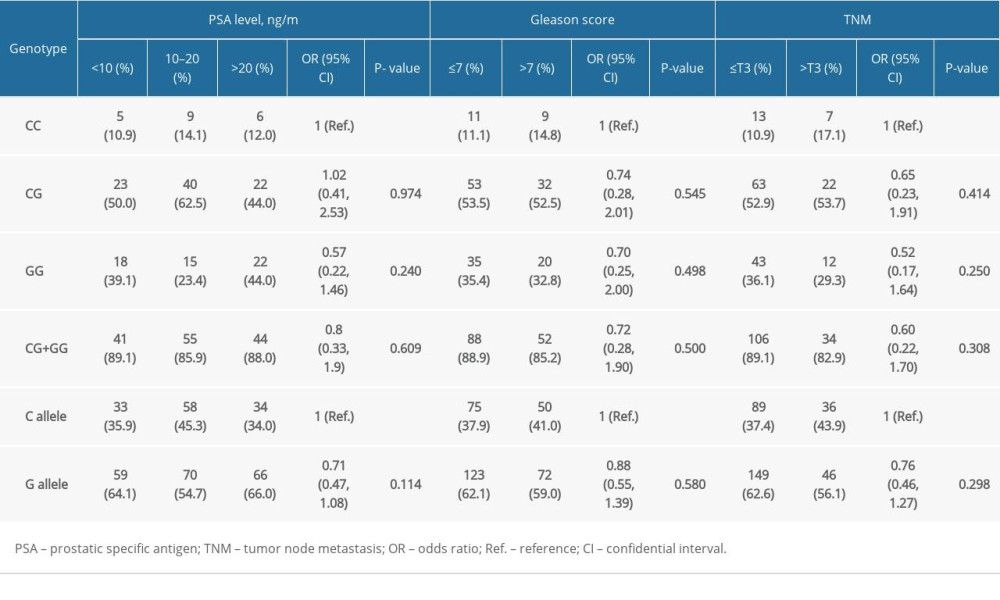 Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis.
Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis.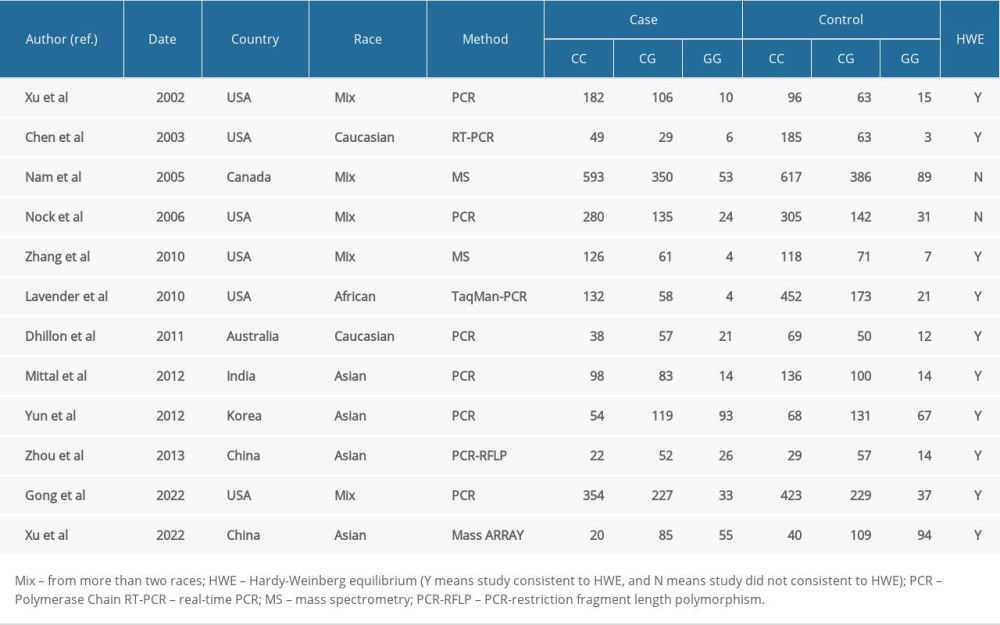 Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk.
Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk.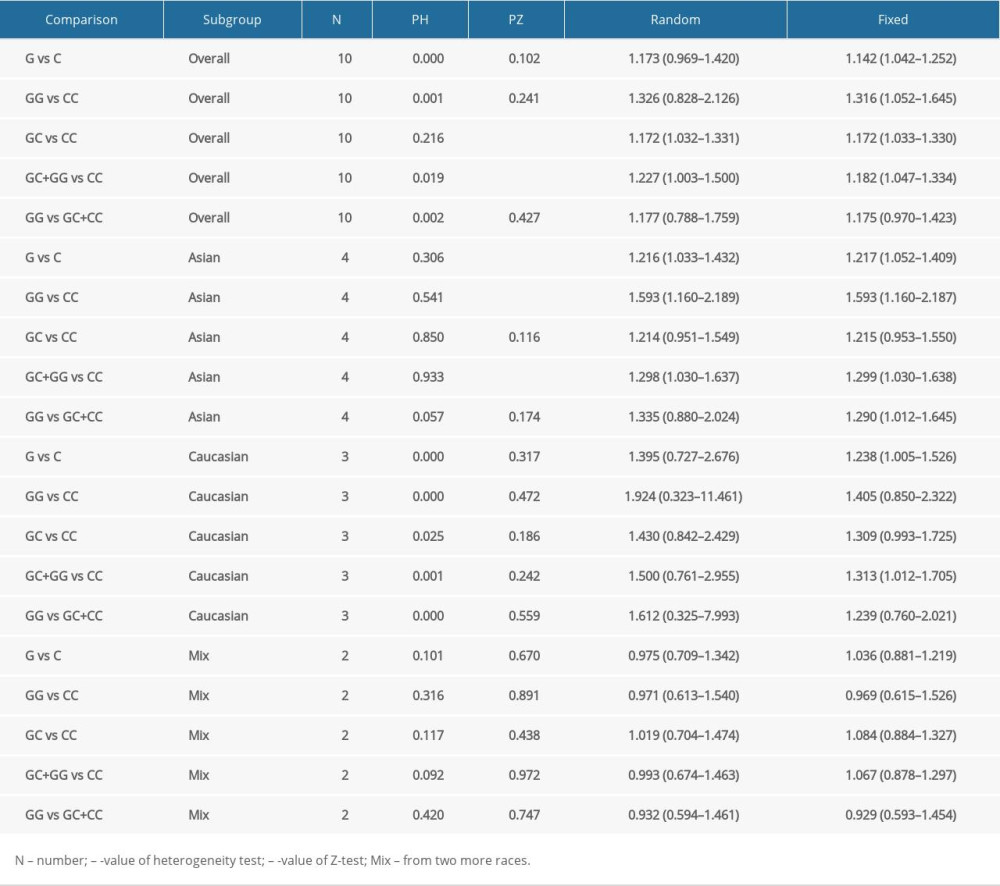 Supplementary Table 1. The NOS scores of the included eleven studies.
Supplementary Table 1. The NOS scores of the included eleven studies.
References
1. Basu AK, DNA damage, mutagenesis and cancer: Int J Mol Sci, 2018; 19(4); 970
2. Khandrika L, Kumar B, Koul S, Maroni P, Koul HK, Oxidative stress in prostate cancer: Cancer Lett, 2009; 282(2); 125-36
3. Pathak SK, Sharma RA, Steward WP, Oxidative stress and cyclooxygenase activity in prostate carcinogenesis: Targets for chemopreventive strategies: Eur J Cancer, 2005; 41(1); 61-70
4. Sikka SC, Role of oxidative stress response elements and antioxidants in prostate cancer pathobiology and chemoprevention – a mechanistic approach: Curr Med Chem, 2003; 10(24); 2679-92
5. Krokan HE, Bjørås M, Base excision repair: Cold Spring Harb Perspect Biol, 2013; 5(4); a012583
6. Sinha RP, Häder DP, UV-induced DNA damage and repair: A review: Photochem Photobiol Sci, 2002; 1(4); 225-36
7. D’Augustin O, Huet S, Campalans A, Radicella JP, Lost in the crowd: How does human 8-oxoguanine DNA glycosylase 1 (OGG1) find 8-oxoguanine in the genome?: Int J Mol Sci, 2020; 21(21); 8360
8. Ba X, Boldogh I, 8-oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions: Redox Biol, 2018; 14; 669-78
9. Ba X, Aguilera-Aguirre L, Rashid QT, The role of 8-oxoguanine DNA glycosylase-1 in inflammation: Int J Mol Sci, 2014; 15(9); 16975-97
10. Wei B, Zhou Y, Xu Z, The effect of hOGG1 Ser326Cys polymorphism on cancer risk: Evidence from a meta-analysis: PLoS One, 2011; 6(11); e27545
11. Kang SW, Kim SK, Park HJ, Chung JH, Ban JY, Human 8-oxoguanine DNA glycosylase gene polymorphism (Ser326Cys) and cancer risk: Updated meta-analysis: Oncotarget, 2017; 8(27); 44761-75
12. Karahalil B, Engin AB, Coşkun E, Could 8-oxoguanine DNA glycosylase 1 Ser326Cys polymorphism be a biomarker of susceptibility in cancer?: Toxicol Ind Health, 2014; 30(9); 814-25
13. Xu J, Zheng SL, Turner A, Associations between hOGG1 sequence variants and prostate cancer susceptibility: Cancer Res, 2002; 62(8); 2253-57
14. Chen L, Elahi A, Pow-Sang J, Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer: J Urol, 2003; 170(6 Pt 1); 2471-74
15. Zhang J, Dhakal IB, Greene G, Polymorphisms in hOGG1 and XRCC1 and risk of prostate cancer: effects modified by plasma antioxidants: Urology, 2010; 75(4); 779-85
16. Lavender NA, Komolafe OO, Benford M, No association between variant DNA repair genes and prostate cancer risk among men of African descent: Prostate, 2010; 70(2); 113-19
17. Dhillon VS, Yeoh E, Fenech M, DNA repair gene polymorphisms and prostate cancer risk in South Australia – results of a pilot study: Urol Oncol, 2011; 29(6); 641-46
18. Mittal RD, Mandal RK, Gangwar R, Base excision repair pathway genes polymorphism in prostate and bladder cancer risk in North Indian population: Mech Ageing Dev, 2012; 133(4); 127-32
19. Mandal RK, Gangwar R, Kapoor R, Mittal RD, Polymorphisms in base-excision & nucleotide-excision repair genes & prostate cancer risk in north Indian population: Indian J Med Res, 2012; 135(1); 64-71
20. Nam RK, Zhang WW, Jewett MA, The use of genetic markers to determine risk for prostate cancer at prostate biopsy: Clin Cancer Res, 2005; 11(23); 8391-97
21. Nock NL, Cicek MS, Li L, Polymorphisms in estrogen bioactivation, detoxification and oxidative DNA base excision repair genes and prostate cancer risk: Carcinogenesis, 2006; 27(9); 1842-48
22. Yun SJ, Ha YS, Chae Y, The hOGG1 mutant genotype is associated with prostate cancer susceptibility and aggressive clinicopathological characteristics in the Korean population: Ann Oncol, 2012; 23(2); 401-5
23. Zhou C, Xie LP, Lin YW, Susceptibility of XPD and hOGG1 genetic variants to prostate cancer: Biomed Rep, 2013; 1(4); 679-83
24. Gong Z, Platek ME, Till C, Associations between polymorphisms in genes related to oxidative stress and DNA repair, interactions with serum antioxidants, and prostate cancer risk: Results from the prostate cancer prevention trial: Front Oncol, 2021; 11; 808715
25. Feng Y, Yao S, Pu Z, Identification of new tumor-related gene mutations in chinese gastrointestinal stromal tumors: Front Cell Dev Biol, 2021; 9; 764275
26. Wang L, Liu W, Jiang W, A miRNA binding site single-nucleotide polymorphism in the 3′-UTR region of the IL23R gene is associated with breast cancer: PLoS One, 2012; 7(12); e49823
27. Liberati A, Altman DG, Tetzlaff J, The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration: PLoS Med, 2009; 6(7); e1000100
28. Stang A, Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses: Eur J Epidemiol, 2010; 25(9); 603-5
29. Yamane A, Kohno T, Ito K, Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo: Carcinogenesis, 2004; 25(9); 1689-94
30. Zhu S, Zhang H, Tang Y, Wang J, Polymorphisms in XPD and hOGG1 and prostate cancer risk: A meta-analysis: Urol Int, 2012; 89(2); 233-40
31. Zhang H, Xu Y, Zhang Z, Li L, The hOGG1 Ser326Cys polymorphism and prostate cancer risk: A meta-analysis of 2584 cases and 3234 controls: BMC Cancer, 2011; 11; 391
32. Chen Y, Li J, Li T, Mo Z, hOGG1 C1245G gene polymorphism associated with prostate cancer: A meta-analysis: Int J Biol Markers, 2015; 30(2); e161-68
33. Agalliu I, Kwon EM, Salinas CA, Genetic variation in DNA repair genes and prostate cancer risk: Results from a population-based study: Cancer Causes Control, 2010; 21(2); 289-300
Figures
Tables
 Table 1. Comparison of clinical pathological characteristics between prostate cancer cases and controls.
Table 1. Comparison of clinical pathological characteristics between prostate cancer cases and controls. Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls.
Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls. Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer.
Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer. Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis.
Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis. Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk.
Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk. Table 1. Comparison of clinical pathological characteristics between prostate cancer cases and controls.
Table 1. Comparison of clinical pathological characteristics between prostate cancer cases and controls. Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls.
Table 2. hOGG1 (rs1052133) gene polymorphism in patients with prostate cancer and controls. Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer.
Table 3. Correlation between hOGG1 (rs1052133) genotypes and different clinicopathological features of prostate cancer. Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis.
Table 4. Major features of eligible case-control studies recruited in the updated meta-analysis. Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk.
Table 5. Summary risk estimations for the relationship of the hOGG1 polymorphism and prostate cancer risk. Supplementary Table 1. The NOS scores of the included eleven studies.
Supplementary Table 1. The NOS scores of the included eleven studies. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952