25 July 2022: Database Analysis
Integrating Network Pharmacology, Molecular Docking, and Experimental Validation to Investigate the Mechanism of (−)-Guaiol Against Lung Adenocarcinoma
Yaoying Zeng1E, Yanbin Pan1B, Bo Zhang1C, Yingbin Luo1D, Jianhui Tian1AF, Yuli Wang1CD, Xudong Ju2BF, Jianchun Wu1A*, Yan Li1ADOI: 10.12659/MSM.937131
Med Sci Monit 2022; 28:e937131
Abstract
BACKGROUND: Lung adenocarcinoma (LUAD) is the most common type of lung cancer, which poses a serious threat to human life and health. -(-)Guaiol, an effective ingredient of many medicinal herbs, has been shown to have a high potential for tumor interference and suppression. However, knowledge of pharmacological mechanisms is still lacking adequate identification or interpretation.
MATERIAL AND METHODS: The genes of LUAD patients collected from TCGA were analyzed using limma and WGCNA. In addition, targets of (-)-Guaiol treating LUAD were selected through a prediction network. Venn analysis was then used to visualize the overlapping genes, which were further condensed using the PPI network. GO and KEGG analyses were performed sequentially, and the essential targets were evaluated and validated using molecular docking. In addition, cell-based verification, including the CCK-8 assay, cell death assessment, apoptosis analysis, and western blot, was performed to determine the mechanism of action of (-)-Guaiol.
RESULTS: The genes included 959 differentially-expressed genes, 6075 highly-correlated genes, and 480 drug-target genes. Through multivariate analysis, 23 hub genes were identified and functional enrichment analyses revealed that the PI3K/Akt signaling pathway was the most significant. Experiment results showed that -(-)Guaiol can inhibit LUAD cell growth and induce apoptosis. Additional evidence suggested that the PI3K/Akt signaling pathway established an inseparable role in the antitumor processes of -(-)Guaiol, which is consistent with network pharmacology results.
CONCLUSIONS: Our results show that the effect of (-)-Guaiol in LUAD treatment involves the PI3K/Akt signaling pathway, providing a useful reference and medicinal value in the treatment of LUAD.
Keywords: Lung Neoplasms, Pharmacology, Signal Transduction, Adenocarcinoma of Lung, Computational Biology, Gene Expression Profiling, Gene Expression Regulation, Neoplastic, Humans, Molecular Docking Simulation, Network Pharmacology, Phosphatidylinositol 3-Kinases, Proto-Oncogene Proteins c-akt, Sesquiterpenes, Guaiane
Background
According to the latest survey of American Cancer Society [1], lung cancer is still the leading cause of cancer-related death for both men and women (22% for men and 22% for women), with a high rate of metastases. This malignant disease, which has high morbidity and mortality rates, is classified into 2 subtypes: small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC). Lung adenocarcinoma (LUAD), a subtype of NSCLC, is the most common subtype of lung cancer, accounting for 40% of all cases [2]. LUAD treatment differs depending on the stage of the disease. Surgical resection or radiotherapy is considered at the early stage [3]. Patients with advanced-stage disease are treated with chemotherapeutic medications such as platinum agents, taxanes, and therapies that target oncogenetic mutation drivers [4]. Unfortunately, the clinical benefit of conventional therapy is limited by toxicity and acquired resistance, causing recurrence and distant metastasis. Patients are eventually susceptible to disease progression, with a poor prognosis. From 2010 to 2016, the 5-year survival rate for metastatic LUAD was only 6% [1]. It is essential to develop new drugs for combination therapy, which is considered a more effective and advanced treatment for overcoming resistance rather than monotherapies.
-(−)Guaiol, which had distinct antibacterial activity, is a natural compound found in medicinal plants. The formula of -(−)Guaiol is C15H26O, and the molecular weight is 222.37g/mol [5]. This sesquiterpene alcohol with the guaiane skeleton has been used for thousands of years as a traditional Chinese medicine and is the main active ingredient found in
In this study, a network was built using network pharmacology, weighted gene co-expression network analysis (WGCNA), and a system bioinformatics approach, which was then validated using molecular docking and experiments. The main goal of the current study was to determine the anticancer effect and molecular mechanism of -(−)Guaiol against LUAD, which complements those of earlier studies. A general study flow chart is shown in Figure 1.
Material and Methods
IDENTIFICATION OF DIFFERENTIALLY-EXPRESSED GENES:
The level 3 RNA sequencing (RNA-seq) profiles were downloaded from the The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) by searching the following keywords “bronchus and lung”, “adenomas and adenocarcinomas”, and “TCGA-LUAD”, consisting of 375 LUAD tissues and 32 adjacent normal tissues transcriptome profiles. After processing the available data with the limma [19] (v: 3.42.2) R package, differentially-expressed genes (DEGs) between tumor and normal tissues were specifically identified in the TCGA cohort, with a false-discovery rate of 0.05.
WGCNA MODULE CONSTRUCTION:
Additionally, WGCNA, a systems biology modality for distinguishing the correlation patterns among genes across microarray samples [20], was used to screen the highly-correlated genes from RNA-seq profiles for summarizing such clusters and correlating modules to external sample traits. To create a scale-free network, the soft-thresholding parameter was defined as=6 (scale-free R2=0.85) using the soft function pickSoftThreshold. Integrating 2 systems biology approaches increased the discrimination ability of candidate genes selection.
PREDICTION OF THE TARGETS OF (−)-GUAIOL:
The study used PubChem [21] (https://pubchem.ncbi.nlm.nih.gov/), the world’s largest chemical information collection website, to search for the SMILES structural formula of (−)-Guaiol. The obtained structure was then fed into ChemMapper [22] (http://lilab.ecust.edu.cn/chemmapper/) to predict corresponding targets. The names of the corresponding targets were normalized in the Uniprot [23] database (https://www.uniprot.org/). Finally, using Cytoscape (v: 3.7.2) software, all attribute data were combined and the “disease-drug-target” network was visualized.
CO-EXPRESSION NETWORK CONSTRUCTION AND SCREENING OF HUB GENES:
After thoroughly preparing for data collection, a co-expression network was created, which connected 3 major gene cohorts (DEGs, WGCNA module genes, and (−)-Guaiol target genes). The network was displayed by a Venn diagram created with the R package VennDiagram [24] (v: 1.6.20). Following that, the co-expression genes were entered into Cytoscape (v: 3.7.2) software for protein–protein interactions (PPI) network construction. They were processed using “Biogenet” [25] (v: 3.0.0), an application for visualizing the relationships between multiple biomolecules, and CytoNCA [26] (v: 2.1.6) for topological analysis. In the network, only eligible genes can be selected. The selection criteria were set as: Interaction Degree (DC) >50 and Betweenness (BC) >100. After double standard screening, the genes that met the selection criteria were ultimately enrolled as key biological targets in the subsequent research.
ENRICHMENT ANALYSIS:
System functional enrichment analyses were performed to elucidate the biological functions and signaling pathways associated with (−)-Guaiol against LUAD to gain more insights into the regulation of selected genes. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) are knowledge-base resources. GO provides information on biological processes (BPs), cellular components (CCs), and molecular functions (MFs), while KEGG connects genomic information with higher-order functional intelligence [27,28]. GO and KEGG pathway analyses with a P value of 0.05 were performed using 3 R packages: clusterProfiler [29] (v: 3.14.3), org.Hs.eg.db (v: 3.10.0), and enrichplot (v: 1.6.1). The R package ggplot2 (v: 3.2.1). were used to create the bubble diagrams for the GO enrichment and KEGG pathway analyses.
MOLECULAR DOCKING:
The current study constructed a KEGG pathway-target relation network based on the enriched pathways with related target proteins, with only the top-ranked pathways enrolled. In addition, Cytoscape (v: 3.7.2) was used for topological analysis with the “Network Analyzer” tools. The analysis revealed that targets with a high degree score were of interest and may participate in the molecular docking analysis. Molecular docking, an in silico structure-based approach that allows the identification of active ingredients of therapeutic interest [30], was used to estimate the molecular interactions between targets and (−)-Guaiol. In addition, the chemical formulas and 3D structures of the samples, CDK2 (PDB ID: 1B39: 2.1Å), FN1 (PDB ID: 2HAZ: 1.7Å), HSP90AA1 (PDB ID: 5NJX: 2.49Å), and HSP90AB1 (PDB ID: 1UYM: 2.45Å), were obtained from the PDB (https://www.rcsb.org/) and PubChem [31] (https://pubchem.ncbi.nlm.nih.gov/) databases. To improve the quality of molecular docking, AutoDockTools (1.5.6) was used to add charge and display rotatable keys to the compound structures, and Pymol was used to modify the protein crystal structures corresponding to the core target genes by removing water molecules and hetero-molecules. Moreover, the AutoDockTools (1.5.6) were applied to add hydrogen atoms and charge operations. It is necessary to transform the protein format from “pdb” to “pdbqt” format and search for active pockets via AutoDock software. After completing the preparation of the ligand (the compound) and receptors (the proteins corresponding to the core target genes), docking operations with AutoDock Vina could finally be initiated. The binding affinity assisted in measuring the conformational stability as a reference.
CHEMICALS AND REAGENTS:
(−)-Guaiol was commercially obtained from Sigma (448575, St. Louis, MO, USA). DMEM medium was purchased from KeyGEN BioTECH (KGM12800-500, Jiangsu, China). Fetal bovine serum (FBS) was provided by Gibco Life Technologies (10099-141, Grand Island, NY, USA). The reagent Z-VAD-FMK (Z-VAD) was obtained from Selleck (S7023, Shanghai, China). Annexin V-FITC/PI Apoptosis Detection Kits were purchased from KeyGEN BioTECH (KGA105-KGA108, Jiangsu, China). Cell Counting Kit-8 was purchased from YEASEN (40203ES76, Shanghai, China). LY294002 was purchased from MedChemExpress (HY-10108, Monmouth Junction, New Jersey, USA). SYTOX Green was purchased from Invitrogen (S7020, Carlsbad, California, USA). The following antibodies – Bcl-2(15071, 1: 1000), BAX (5023, 1: 1000), β-actin (4970, 1: 1000), p-Akt (4060, 1: 1000), Akt (2920, 1: 1000), PI3K (4255, 1: 1000), p-PI3K (4249, 1: 1000), and IgG secondary antibody (10700, 1: 2000) – were purchased from Cell Signaling Technology (Boston, Massachusetts, USA).
CELL CULTURE AND DRUG ADMINISTRATION:
The LUAD cells A549 and Calu-1 and normal lung cells BEAS-2B were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin streptomycin in a humidified atmosphere with 5% CO2 at 37°C. (−)-Guaiol was diluted in methanol at a primary stock concentration of 10 mM. The control group was DMEM with 10% FBS and methanol.
CELL SURVIVAL ASSAY:
Cell survival was detected with a CCK-8 assay kit. Approximately 5000 cells were seeded into a 96-well culture plate, and after 24 h, the cells were exposed to various doses of (−)-Guaiol (60 μM or150 μM) or Z-VAD (20 μM). The solution was then removed, and all wells were washed twice with PBS before adding 10 μl CCK-8 solution to the wells for incubation. After waiting 1h, a Multiplate Reader (Bio Tek, United States) was used to measure the OD value at 450 nm. The cell survival rate (%) was calculated as ([experimental group-blank group]/[control group-blank group]) 100% using the average OD value of 3 wells.
SYTOX Green staining assay was performed according to the manufacturer’s protocol, visualizing cell death through imaging applications. The working concentration was 100 nM and experiments were carried out 3 times each.
APOPTOSIS ANALYSIS:
The cells were inoculated into 6-well plates and each well contained 2 mL of media with 10% FBS and approximately 2×105 cells. They were cultured for 24 h at 37°C and 5% CO2, then treated by (−)-Guaiol (60 μM or 120 μM) for 24 h. The cells were collected after drug treatment. We added binding buffer (500 μL), Annexin V-FITC (5 μL), and propidium iodide (5 μL) to each tube, followed by incubation at room temperature in the dark for 10 min. Finally, a fluorescence microscope was used to observe the cell morphology and flow cytometry was used to detect the apoptosis rate. The experiments were carried out 3 times each.
WESTERN BLOT ANALYSIS:
After treatment, the cells were collected and washed twice with PBS, then added to cell lysis buffer with protease and phosphatase inhibitors for 30 min at 4°C. The whole protein was quantified with a bicinchoninic acid kit (Beyotime, China). Samples were obtained after boiling within the 1% SDS loading buffer at 100°C for 8 min. Afterward, 20 μg protein was subjected to SDS-PAGE and subsequently transferred to a PVDF membrane. After immersing the membrane in BCA for blocking, the samples were incubated overnight with the following primary antibodies – β-actin, p-Akt (Ser473), Akt, PI3K, p-PI3K (110α), BAX, and Bcl-2 – diluted 1: 1000. Next, we used an anti-rabbit IgG secondary antibody (1: 2000) to conjugate each primary antibody. Finally, the samples mixed with ECL (Tanon, Shanghai, China) solution were exposed and recorded by chemiluminescence reagent. The gray values of the bands, which were calculated automatically using Image J software, were normalized to β-actin as the internal control. The relative protein levels were calculated by normalizing the densitometry results of the experimental group to those of the control group. One-way ANOVA was used to statistically analyze the significance. The experiments were carried out 3 times each.
DATA ANALYSIS:
All results were obtained from experiments independently repeated 3 times. The continuous variables of normal distribution or nearly normal distribution were displayed as average value±standard deviation. After homogeneity of variance test, one-way ANOVA was used for those meeting data requirements in SPSS 25.0. or GraphPad Prism 8.0.
Results
CONSTRUCTION OF DEGS AND WGCNA MODULE IN TCGA COHORT:
There were 15 208 RNA-seq genes of LUAD downloaded from the TCGA cohort, with 959 genes pertaining to DEGs and 6075 genes pertaining to WGCNA key modules. The 959 DEGs were described by the volcano plot and heat map, using the R package ggplot2 [32] (v: 3.2.1) for visualization (Figure 2A, 2B). The 6075 highly-related genes were classified into 15 color modules and are presented in Figure 2C, 2D.
TARGET PREDICTION AND ANALYSIS OF (−)-GUAIOL:
A prediction network integrating targets of (−)-Guaiol against LUAD was obtained during this process. Figure 3 depicts the “disease-drug-target” network. As a result, 480 targets were identified and are represented by the green ovals in the plot; thus far, all of the target genes have been collected under the current conditions. The file containing total gene (DEGs, WGCNA module genes and drug target genes) information was included as Supplementary Table 1.
IDENTIFICATION OF HUB GENES:
The genes were then optimized and reduced to a few hub genes in the following step of the workflow. The selected co-expression genes among DEGs, WGCNA module genes, and (−)-Guaiol target genes were presented in a Venn diagram (Figure 4A), and only 17 genes were filtered out in the plot. The 17 co-expression genes were then analyzed by PPI network-to-topology analysis. The PPI network was established for topology feature analysis with the Cytoscape software platform based on degree and betweenness parameters for further investigation. There were 23 genes that met the degree >50 and betweenness >100 thresholds (Figure 4B), which meant these genes played an essential role in (−)-Guaiol treatment of LUAD. The specific gene information on 17 identified genes and 23 hub genes is shown in Supplementary Tables 2 and 3.
GO AND KEGG ENRICHMENT ANALYSIS:
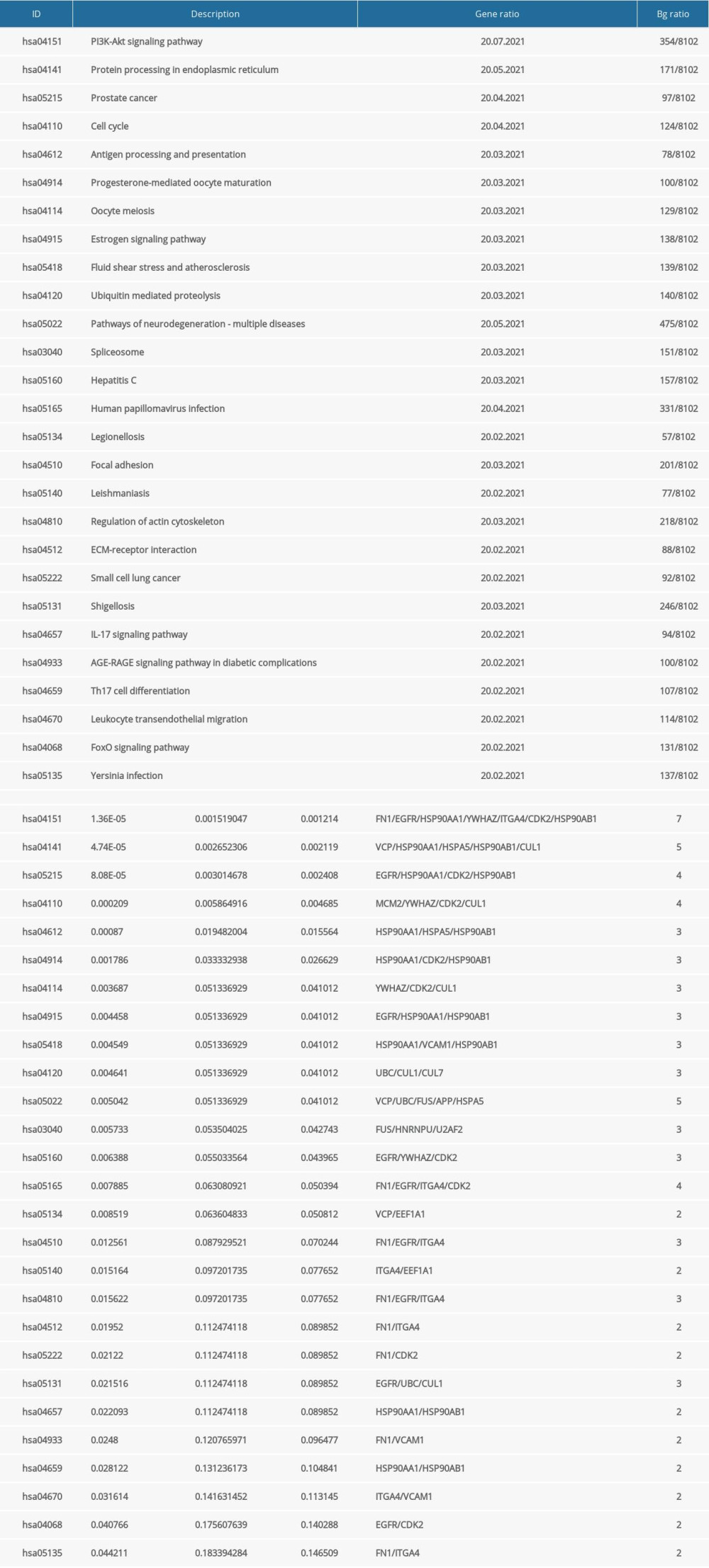
The 23 genes that emerged were used in subsequent functional enrichment analyses. The GO and KEGG enrichment analyses identified a number of related biological activities that were statistically significant (Figure 5). In total, 883 significant GO terms and 27 significant pathways were enriched (P<0.05) (Supplementary Table 4). The BPs aspect changes were clearly associated with 732 functional categories, including DNA metabolic process regulation, developmental cell growth regulation, protein binding regulation, and positive cell cycle regulation. Furthermore, there were 81 terms in the CCs analysis aspect, primarily vesicle lumen, secretory granule lumen, apical plasma membrane, and membrane microdomain. There were also 70 terms in MFs, such as ubiquitin protein ligase binding, cell adhesion molecule binding, MHC class II protein complex binding, and ATPase activity. The KEGG pathway enrichments revealed that the identified targets were mainly enriched in 27 pathways, including the hosphatidylinositol3-kinase/ProteinKinase-B (PI3K/Akt) signaling pathway, protein processing in the endoplasmic reticulum, cell cycle, and antigen processing and presentation. Of note, the PI3K-Akt signaling pathway was significant, which is listed at the top of the chart.
KEGG PATHWAY-TARGET NETWORK AND MOLECULAR DOCKING:
The pathway-target network for (−)-Guaiol and LUAD, which shows 19 targets and 20 pathways, was built to reveal potential mechanisms of action. Based on the calculated degree, each node pertaining to the pathway or target was displayed in an ordered arrangement (Figure 6). According to the network, the top 5 candidate targets (CDK2, EGFR, FN1, HSP90AA1, HSP90AB1) were investigated further for their interaction with (−)-Guaiol; notably, these genes were also involved in the PI3K/Akt signaling pathway in KEGG analysis. Each target was docked with the (−)-Guaiol 3D structure after a series of modifications. As shown in Figure 7, CDK2, FN1, HSP90AA1 and HSP90AB1 displayed a favorable binding affinity, with ideal conjunction in general. The docking score in binding affinity represents the cohesiveness of the models. It is clear that the lower binding affinity score was preferred.
(−)-GUAIOL CAN SUPPRESS LUAD CELLS AND INDUCE APOPTOSIS:
The CCK-8 assay kit was used to assess the cytotoxicity of (−)-Guaiol in A549 cells and Calu-1 cells, taking normal lung cells BEAS-2B as a control. The survival rates of A549 and Calu-1 were concentration-dependently decreased when treated with different concentrations of (−)-Guaiol, as suggested by the CCK-8 assay (Figure 8A). According to the results of GraphPad Prism, (−)-Guaiol dramatically inhibited growth of A549 cells and Calu-1 cells, with IC50 values of 75.7 μM and 92.1 μM, respectively, whereas its IC50 value for BEAS-2B cells was 322.3 μM. To investigate the apoptosis action of (−)-Guaiol, the appropriate concentration (75 μM) of (−)-Guaiol and the apoptosis inhibitor Z-VAD were used in the cell death assays. When compared to the (−)-Guaiol single group, Z-VAD significantly reversed cell viability in A549 (p=0.048) and Calu-1 cells (P=0.033) (Figure 8B). Similarly, the SYTOX Green staining assay showed that (−)-Guaiol damaged LUAD cells. Under fluorescence microscopy, the exceptional brightness of the signal produced by the (−)-Guaiol group indicated more dead cells, whereas Z-VAD weakened the fluorescence intensity, counteracting the effect of (−)-Guaiol, which was consistent with the CCK-8 assay results (Figure 8C).
(−)-GUAIOL INCREASED EARLY APOPTOSIS AND ALTERED EXPRESSION OF RELATED PROTEINS:
The Annexin V-FITC/PI dual-staining assay was used for quantitative analysis to gain more insight into the apoptosis induced by (−)-Guaiol. As shown in Figure 9A, the early apoptosis of the control group, 60 μM (−)-Guaiol group, and 120 μM (−)-Guaiol group for A549 was 17.75%, 25.9%, and 35.7%, respectively, and for Calu-1 it was 1.07%, 10.9%, and 13.9%, respectively. (−)-Guaiol induced apoptosis by increasing early apoptosis, as demonstrated by the detected green fluorescence indicating PS in apoptotic cells with low red fluorescence in the image, and the proportion of cells with a higher level of green and lower level of red fluorescence was higher in the flow cytometry analysis (Figure 9B). According to western blot analysis, (−)-Guaiol inhibited the expression of the anti-apoptotic protein Bcl-2 (p<0.001) while increasing the expression of the pro-apoptotic protein BAX (P=0.028) (Figure 9C).
(−)-GUAIOL REGULATES THE EXPRESSION OF RELATED PROTEINS IN PI3K/AKT SIGNALING PATHWAY:
The PI3K/Akt signaling pathway, which includes classical proteins PI3K, phosphorylated PI3K, Akt, and phosphorylated Akt, was found to be highly likely to be involved in the mechanisms of (−)-Guaiol’s anti-LUAD activity. To validate the conclusion, western blotting was used to examine the protein expression of PI3K/Akt signaling in A549 cells treated with (−)-Guaiol (75 μM) and LY294002 (15 μM), a PI3K/Akt signaling pathway inhibitor. The expression of total PI3K and Akt was little changed (P>0.05). Treatment with (−)-Guaiol decreased p-PI3K (P=0.001) and p-Akt (P<0.001) levels and furthered the LY294002-induced decrease in p-PI3K (P<0.001) and p-Akt levels (P=0.004), as expected (Figure 10). These findings suggest that (−)-Guaiol inhibits the PI3K/Akt signaling pathway, which regulates apoptosis in A549 cells, thereby validating the network pharmacology result.
Discussion
Lung cancer is the most prevalent human malignancy worldwide; historically, LUAD became the most common subtype of lung cancer, outranking squamous cell carcinoma (SCC) by 1988 to 2002 [33]. As a result, numerous studies on LUAD treatment have been conducted. Medical research combined with network pharmacology is an appealing approach that is becoming increasingly popular in scientific research fields [34]. In any case, a better understanding of the underlying mechanism is essential for guiding effective and precise therapy.
(−)- Guaiol has shown potent antineoplastic activity, but the molecular mechanism has yet to be fully understood. To clarify the underlying mechanism, a network was constructed based on the significant genes, and through a series of network analyses, pivot targets and pathways were discovered. First, the gene information of LUAD patients was collected using the publicly available database TCGA, which was then processed using computational methods to generate 959 DEGs between tumor and non-tumor tissues, as well as 6075 genes highly correlated with WGCNA. At the same time, network pharmacology identified another 480 (−)-Guaiol target genes. These were extracted and shrunk to 17 co-expression genes, and they eventually yielded 23 hub genes through topological data analysis of PPI. Based on the findings, a GO biological process and KEGG pathway enrichment analyses were performed to investigate the potential mechanisms. However, there are some limitations to our study. As we know, bioinformatics has opened up a new field of pharmacological research that develops computing technology and networks public databases for analysis of biological data. Researchers use network databases for data collection, but some related genes may still have been missed, which is unavoidable. Only strongly correlated and significant genes can be used for subsequent analysis. Bioinformatics analysis provides significant options, although it may not be comprehensive. Based on the result of the enrichment analysis, we mainly focused on the PI3K/Akt signaling pathway, which was the most significant pathway in enrichment analysis.
The study was then completed with validation, which included molecular docking and experiments. The molecular docking study confirmed that the top 5 candidate targets from the KEGG pathway-target network (CDK2, EGFR, FN1, HSP90AA1, HSP90AB1), which also were found to be involved in the PI3K/Akt signaling pathway, had a high affinity for (−)-Guaiol, enhancing the results of network pharmacology. Specifically, Cyclin-dependent kinase 2 (CDK2) plays a vital role in cycle regulation and DNA damage response. In addition, CDK2 induces apoptosis by targeting FOXO1, a pivotal protein in triggering DNA-damage-induced apoptosis following dsDNA breaks [35], but CDK2 acts has opposite effects in cell apoptosis. In KRAS-mutant lung cancer models, inhibiting CDK2 kinase activity causes anaphase catastrophe and apoptosis [36]. The epidermal growth factor receptor (EGFR) is another cell growth regulator that belongs to the receptor tyrosine kinase (RTK) family, stimulating specific downstream signaling pathways such as PI3K/Akt, RAS/RAF/MAPK, and JAK/ATAT [37]. EGFR, which has been identified as an oncogenic driver of NSCLC, is associated with advanced disease stage and poor prognosis [38]. Coincidentally, fibronectin 1(FN1), heat shock protein 90AA1 (HSP90AA1), and heat shock protein 90AB1 (HSP90AB1) are overexpressed in cancer, predicting a poor prognosis [39–41].
Furthermore, a series of experiments were carried out to investigate the PI3K/Akt pathway and effect of (−)-Guaiol in LUAD. The PI3K/Akt pathway, which had the lowest
RTK stimulation and somatic mutations in specific elements pathway elements are the 2 most common mechanisms of PI3K/Akt activation in human cancers [44]. However, somatic mutations and amplifications of the PIK3CA gene, which encodes the class I PI3K p110α, frequently occur in patients with NSCLC. Specifically, somatic mutations in PI3KCA are found in 3% to 10% of squamous cell carcinomas and 0% to 2.7% of adenocarcinomas, while PI3KCA amplification is found in 35% of squamous cell carcinomas and 7% of adenocarcinomas [45,46]. Moreover, laboratory research investigated various lung cancer call lines and tumor tissues, including 86 NSCLC cell lines, 43 SCLC cell lines, 3 extrapulmonary small-cell cancer (ExPuSC) cell lines, and 691 resected NSCLC tumors, showing that 12.8% of cell lines and 19.1% of tumors were associated with PIK3CA mutations or amplification [47]. Furthermore, high phosphorylated Akt expression was found in a significant proportion of NSCLC patients, which influenced PIK3CA mutations or amplification [43,47].
The aberrantly activated PI3K/Akt signaling pathway triggers a variety of molecular mechanisms, resulting in a series of changes in cell metabolism and cell growth. Active Akt has been shown to promote cell cycle progression and inhibit apoptosis via a variety of biological mechanisms, including phosphorylation of intermediate targets (GSK3, FOXO, BAD, and MDM2), inhibition of NFB, and activation of mTOR [43]. The Bcl-2 family, a related downstream pathway component, is well known as an apoptosis regulator. The Bcl-2 family plays 2 roles in apoptotic regulation: anti-apoptotic executor and pro-apoptotic executor [48]. Bcl-2 and Bcl-xl are anti-apoptotic proteins that sequester caspases, a unique set of cysteine proteases that cleave critical cellular proteins to cause apoptotic changes or prevent mitochondrial apoptogenic factors from release into the cytoplasm. Other members of the Bcl-2 family, such as BAX and Bak, are responsible for pro-apoptotic functions such as facilitating caspase release and activation depending on mitochondrial outer-membrane permeabilization (MOMP) [49]. According to the findings, (−)-Guaiol inhibited Bcl-2 expression while increasing BAX expression, resulting in lung cancer cell apoptosis.
(−)-Guaiol was found to kill LUAD cells by inhibiting the PI3K/Akt signaling pathway and inducing cell apoptosis. However, (−)-Guaiol has little influence on normal lung cells. The apoptosis inhibitor Z-VAD was used to investigate the apoptosis effect of (−)-Guaiol in cell death assays. The CCK8 and SYTOX Green staining assay illustrated that the Z-VAD rescued the LUAD cells in (−)-Guaiol treatment, suggesting (−)-Guaiol causes apoptosis in LUAD. The apoptotic cells were mainly early apoptotic cells in the apoptosis analysis. Interestingly, previous research found that (−)-Guaiol activated double-strand breaks (DSBs)-induced cell apoptosis via caspase signaling and targeting RAD51, a key factor in homologous recombination repair [15]. Another study looked into the mechanism of autophagy and discovered that (−)-Guaiol targeted the mTORC1 and mTORC2 signaling pathways to cause autophagic cell death. Notably, the mTORC2-AKT signaling was blocked due to mTOR phosphorylation failure, and the mTORC1 signaling was impaired due to downstream factor inhibition [16]. In light of this, PI3K/Akt is regarded as a common upstream signaling pivot, initiating multiple downstream activities leading to cell death.
Conclusions
The purpose of this study was to assess the effects and mechanism of (−)-Guaiol against LUAD using network pharmacology and bioinformatics. As part of the validation process, molecular docking and experiments were carried out. The results showed that (−)-Guaiol targeted the PI3K/Akt pathway to exert its effects via the apoptotic pathway. These findings support the results of previous research on (−)-Guaiol, which indicate that (−)-Guaiol is a potential candidate for clinical application and promotion. Additional research on (−)-Guaiol, clinical trials, and more elaborate experiments are needed.
Figures
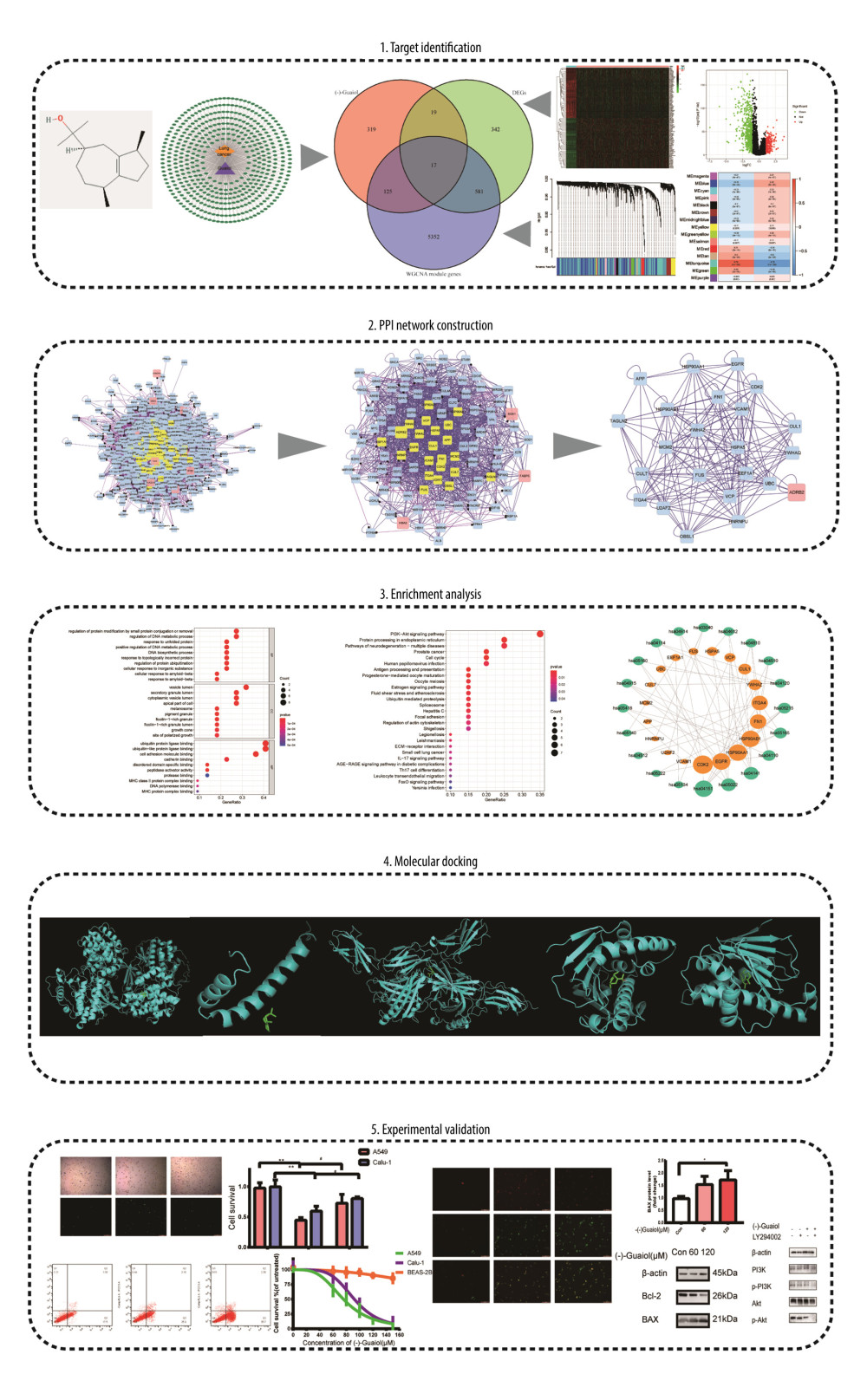 Figure 1. In the flow chart, the systems pharmacology approach was used to determine the mechanism of (−)-Guaiol in the treatment of LUAD by integrating target identification, network construction, enrichment analysis, molecular docking and experimental validations.
Figure 1. In the flow chart, the systems pharmacology approach was used to determine the mechanism of (−)-Guaiol in the treatment of LUAD by integrating target identification, network construction, enrichment analysis, molecular docking and experimental validations. 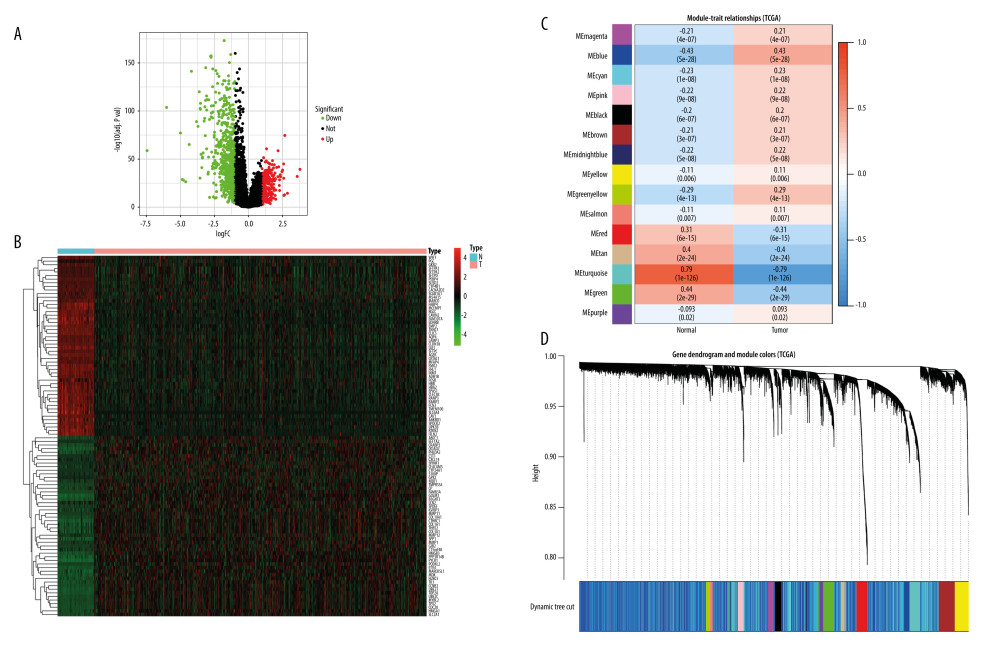 Figure 2. (A) Volcano plot of DEGs in TCGA. (B) Heat map of DEGs in TCGA. The green parts present down-regulated genes, red parts present up-regulated genes, and black parts presents non-differential genes. (C) The cluster dendrogram with different colors of TCGA. (D) Module-trait relationships. Each row corresponds to a color, while each column corresponds to the normal or tumor group.
Figure 2. (A) Volcano plot of DEGs in TCGA. (B) Heat map of DEGs in TCGA. The green parts present down-regulated genes, red parts present up-regulated genes, and black parts presents non-differential genes. (C) The cluster dendrogram with different colors of TCGA. (D) Module-trait relationships. Each row corresponds to a color, while each column corresponds to the normal or tumor group. 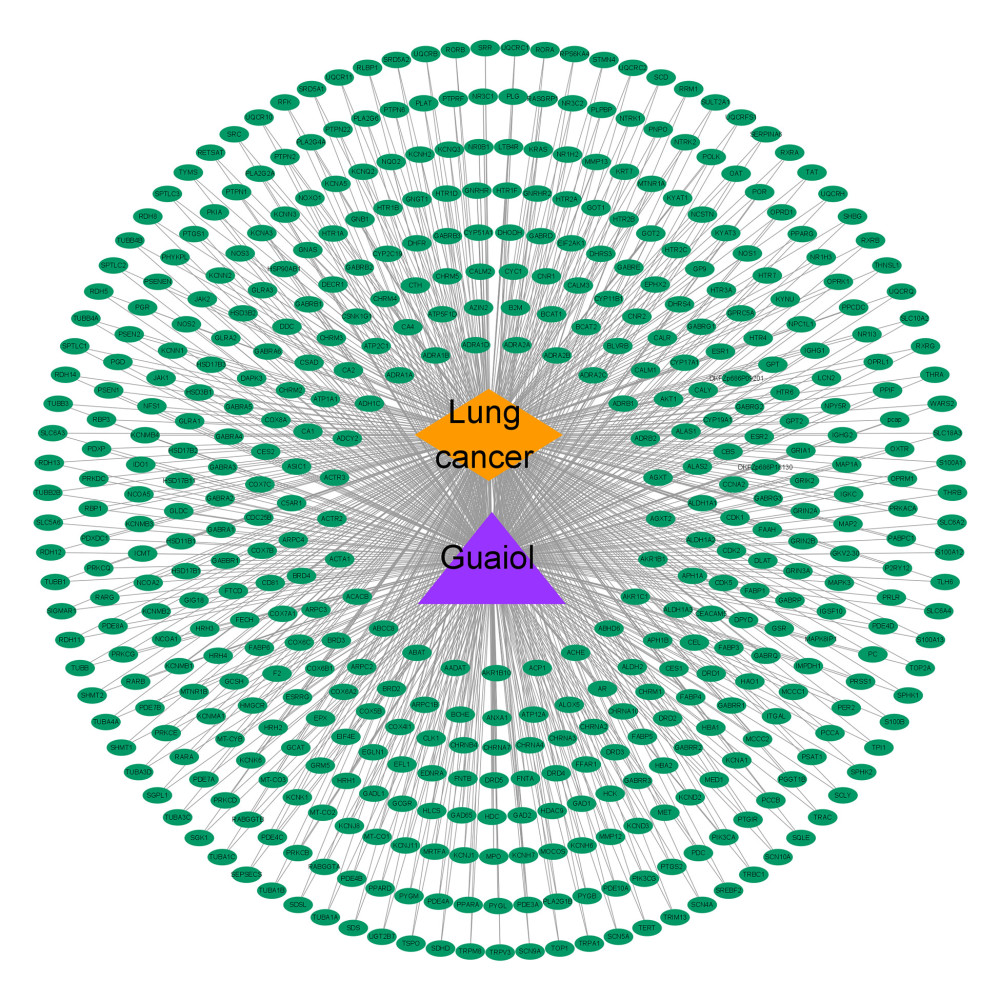 Figure 3. Disease-drug-target network. Purple triangular node presents the drug, orange rhombic node presents disease, and green oval nodes present 480 targets.
Figure 3. Disease-drug-target network. Purple triangular node presents the drug, orange rhombic node presents disease, and green oval nodes present 480 targets. 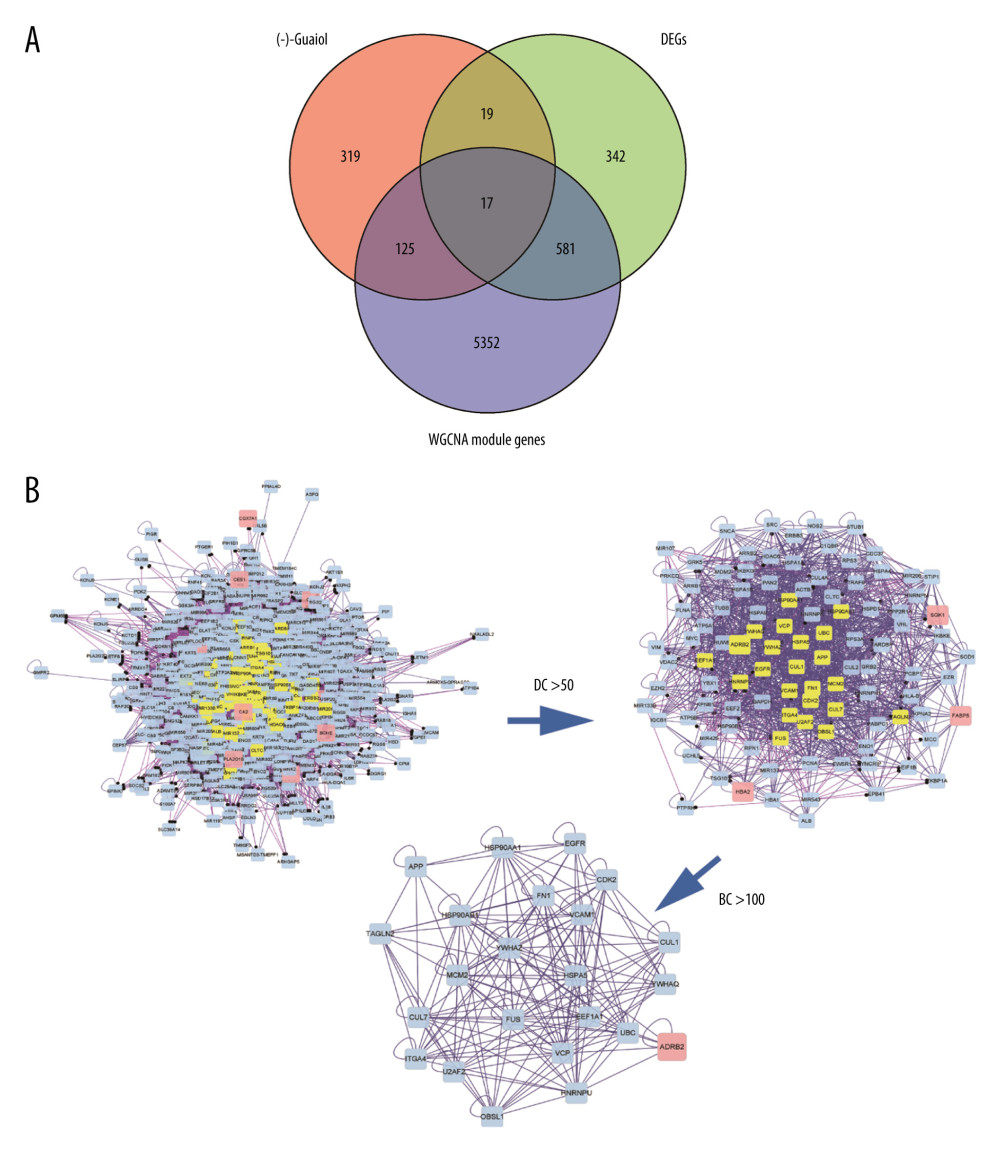 Figure 4. (A) Venn diagram was used to summarize the co-expression genes of DEGs, WGCNA module genes, and (−)-Guaiol target genes. (B) The 23 hub genes were predicted by topological screening of the protein–protein interaction (PPI) network with screening 2 times. DC – degree; BC – betweenness.
Figure 4. (A) Venn diagram was used to summarize the co-expression genes of DEGs, WGCNA module genes, and (−)-Guaiol target genes. (B) The 23 hub genes were predicted by topological screening of the protein–protein interaction (PPI) network with screening 2 times. DC – degree; BC – betweenness. 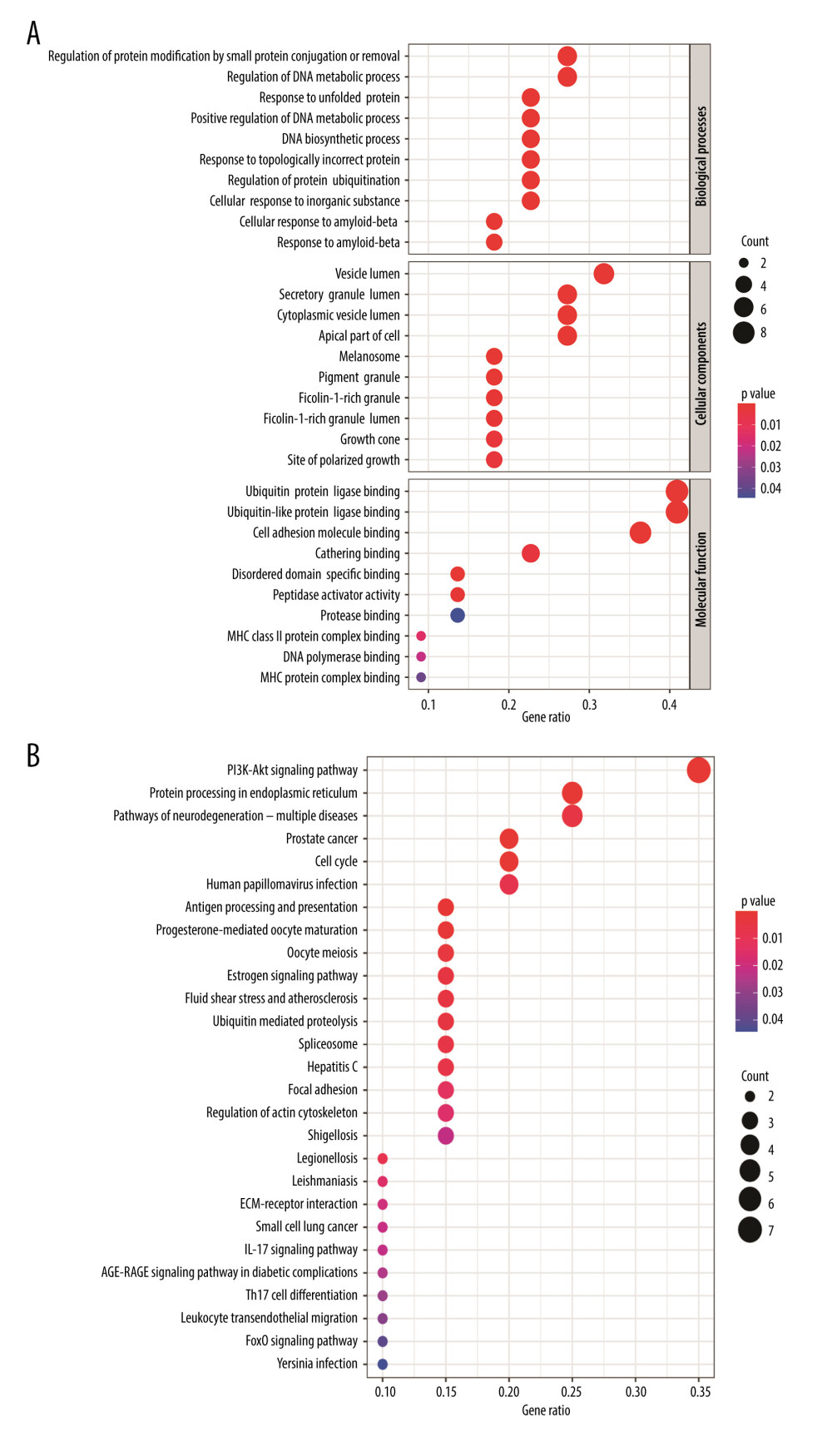 Figure 5. (A) Representative results (P<0.05) of GO enrichment analysis. (B) Representative results (p<0.05) of KEGG enrichment analyses. Bubble size indicates gene count and the color indicates P value.
Figure 5. (A) Representative results (P<0.05) of GO enrichment analysis. (B) Representative results (p<0.05) of KEGG enrichment analyses. Bubble size indicates gene count and the color indicates P value. 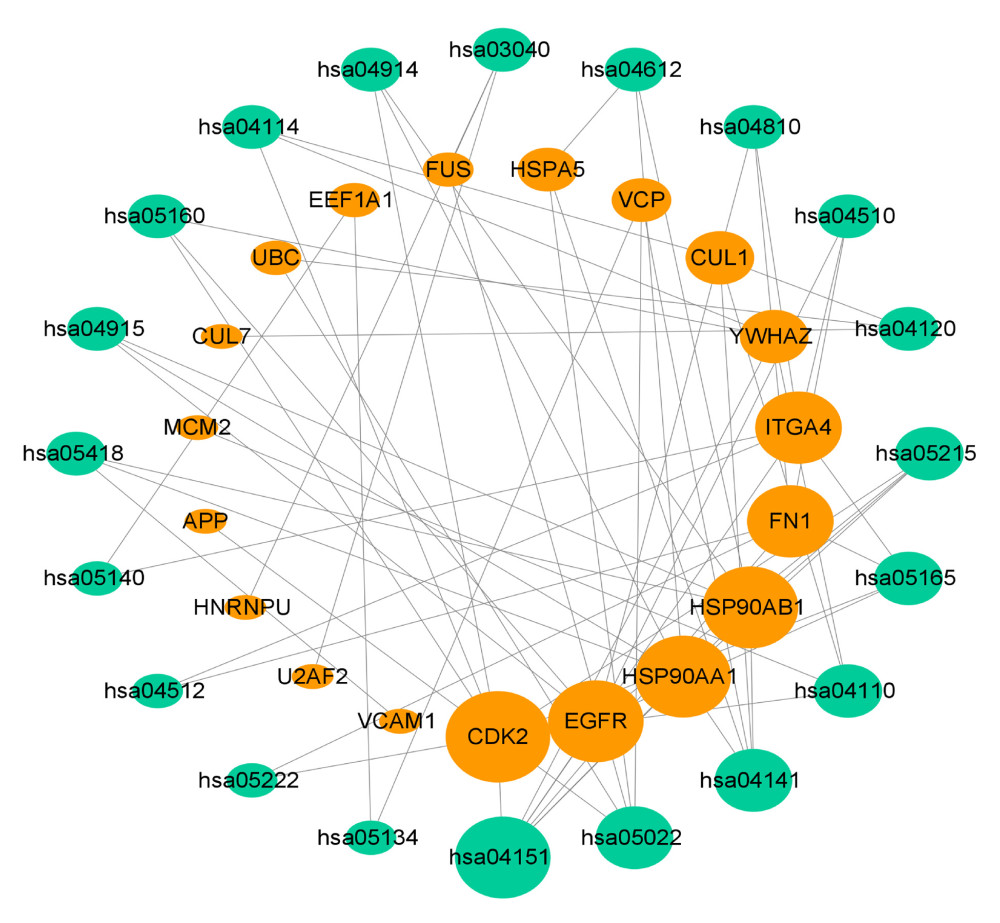 Figure 6. KEGG pathway-target network. The orange round nodes represent the top 19 related targets and the green round nodes represent the top 20 pathways.
Figure 6. KEGG pathway-target network. The orange round nodes represent the top 19 related targets and the green round nodes represent the top 20 pathways. 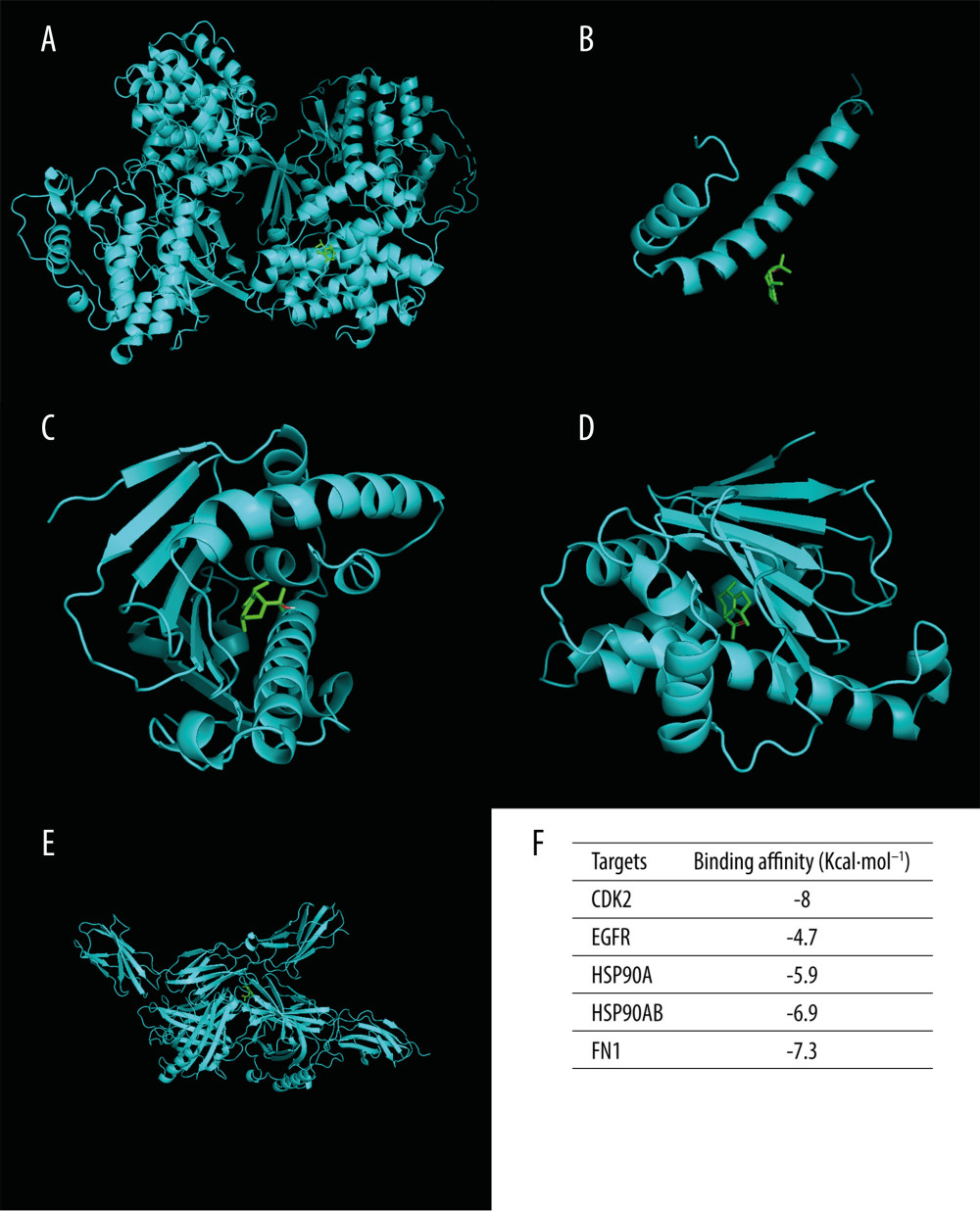 Figure 7. Molecular docking diagrams of (−)-Guaiol with (A) CDK2, (B) EGFR, (C) HSP90AA1, (D) HSP90AB1, and (E) FN1. (F) The binding affinity for the 5 targets docked into the (−)-Guaiol crystal structure.
Figure 7. Molecular docking diagrams of (−)-Guaiol with (A) CDK2, (B) EGFR, (C) HSP90AA1, (D) HSP90AB1, and (E) FN1. (F) The binding affinity for the 5 targets docked into the (−)-Guaiol crystal structure. 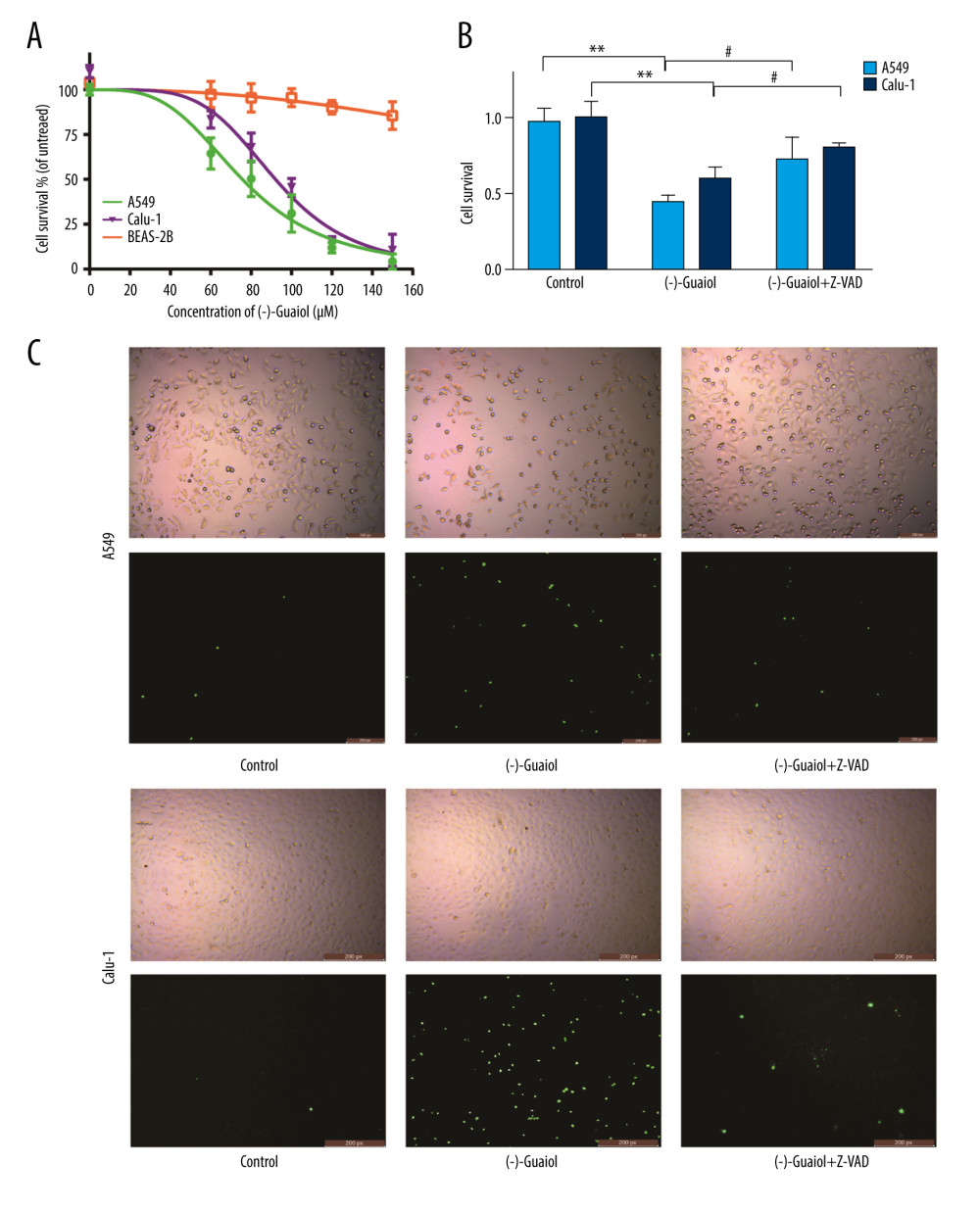 Figure 8. (A) Cell survival rate was detected by performing a CCK-8 assay. The LUAD cells (A549 and Calu-1) and normal lung cells BEAS-2B cells treated with different concentrations of (−)-Guaiol. IC50 values were 75.7 μM for A549 cells, 92.1 μM for H1299 cells, and 322.3 μM for BEAS-2B cells. Data from 3 independent experiments were represented as means±SD. (B) A549 and Calu-1 were treated with (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) companied Z-VAD (20μM) for 24 h. ** P<0.01 vs control in A549. # P<0.05 vs (−)-Guaiol in Calu-1. Data from 3 independent experiments were represented as means±SD. (C) A549 and Calu-1 were treated with or without (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) and Z-VAD (20 μM), and stained with SYTOX Green (100 nM), then observed under a fluorescence microscope.
Figure 8. (A) Cell survival rate was detected by performing a CCK-8 assay. The LUAD cells (A549 and Calu-1) and normal lung cells BEAS-2B cells treated with different concentrations of (−)-Guaiol. IC50 values were 75.7 μM for A549 cells, 92.1 μM for H1299 cells, and 322.3 μM for BEAS-2B cells. Data from 3 independent experiments were represented as means±SD. (B) A549 and Calu-1 were treated with (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) companied Z-VAD (20μM) for 24 h. ** P<0.01 vs control in A549. # P<0.05 vs (−)-Guaiol in Calu-1. Data from 3 independent experiments were represented as means±SD. (C) A549 and Calu-1 were treated with or without (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) and Z-VAD (20 μM), and stained with SYTOX Green (100 nM), then observed under a fluorescence microscope. 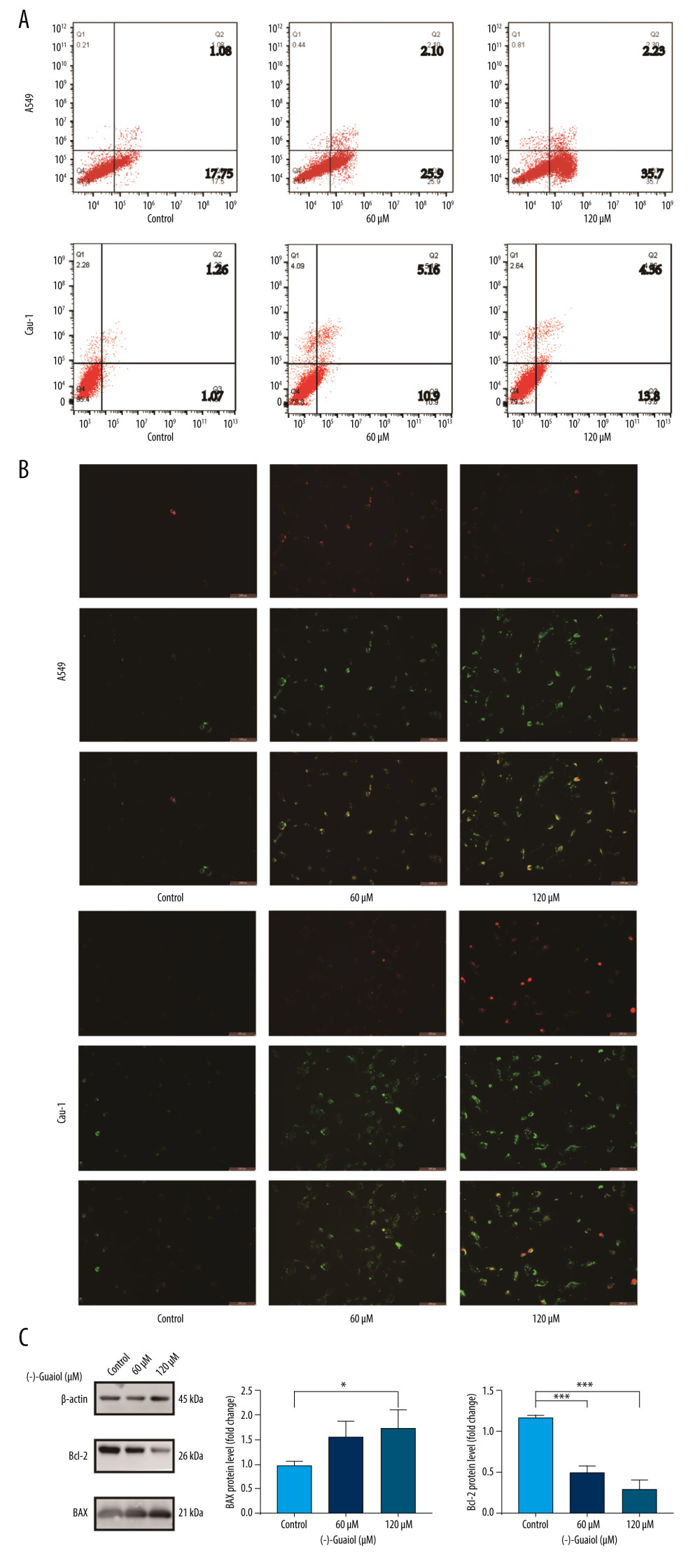 Figure 9. (A) Flow cytometry to detect the apoptosis rate in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (B) Fluorescence microscope was used to observe apoptosis in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (C) Western blot analysis was used to detect the protein expression of BAX and Bcl-2 in A549 treated with (−)-Guaiol (60 μM or 120 μM). The relative BAX and Bcl-2 levels from 3 independent experiments were statistically analyzed using ANOV A test in the quantitative analysis. * P<0.05, *** P<0.001 vs control.
Figure 9. (A) Flow cytometry to detect the apoptosis rate in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (B) Fluorescence microscope was used to observe apoptosis in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (C) Western blot analysis was used to detect the protein expression of BAX and Bcl-2 in A549 treated with (−)-Guaiol (60 μM or 120 μM). The relative BAX and Bcl-2 levels from 3 independent experiments were statistically analyzed using ANOV A test in the quantitative analysis. * P<0.05, *** P<0.001 vs control. 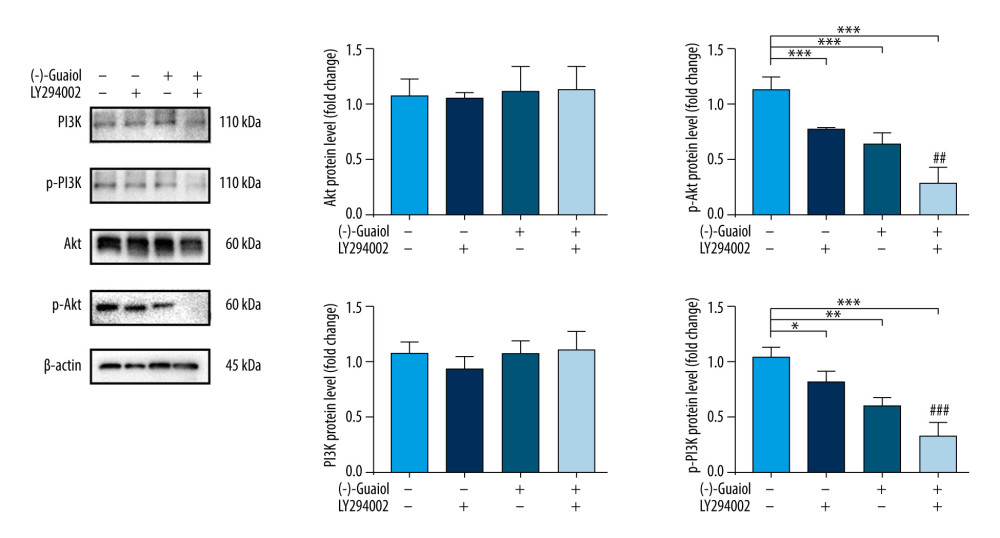 Figure 10. (−)-Guaiol regulates the expression of related proteins in PI3K/Akt Signaling Pathway. Western blot analysis was used to detect the protein expression of PI3K, p-PI3K, Akt, and p-Akt in A549 treated with or without (−)-Guaiol (75 μM) or LY294002 (15 μM). The relative protein levels from 3 independent experiments were statistically analyzed using ANOVA in the quantitative analysis. * P<0.05, ** P<0.01, *** P<0.001 vs control. ## P<0.01, ### P<0.001 vs LY294002.
Figure 10. (−)-Guaiol regulates the expression of related proteins in PI3K/Akt Signaling Pathway. Western blot analysis was used to detect the protein expression of PI3K, p-PI3K, Akt, and p-Akt in A549 treated with or without (−)-Guaiol (75 μM) or LY294002 (15 μM). The relative protein levels from 3 independent experiments were statistically analyzed using ANOVA in the quantitative analysis. * P<0.05, ** P<0.01, *** P<0.001 vs control. ## P<0.01, ### P<0.001 vs LY294002. References
1. Siegel RL, Miller KD, Fuchs HE, Cancer statistics, 2021: Cancer J Clin, 2021; 71(1); 7-33
2. Denisenko TV, Budkevich IN, Zhivotovsky B, Cell death-based treatment of lung adenocarcinoma: Cell Death Dis, 2018; 9(2); 117
3. Duma N, Santana-Davila R, Molina JR, Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment: Mayo Clin Proc, 2019; 94(8); 1623-40
4. Horn L, Advances in the treatment of non-small cell lung cancer: J Natl Compr Canc Netw, 2014; 12; 764-67
5. Kim S, Chen J, Cheng T, PubChem in 2021: New data content and improved web interfaces: Nucleic Acids Res, 2021; 49(D1); D1388-95
6. Shu Z, Pu J, Chen L, Alisma orientale: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine: Am J Chin Med, 2016; 44(2); 227-51
7. Yang M, Luo J, Li K, Determination and pharmacokinetic study of guaiol in rat plasma by gas chromatography-mass spectrometry with selected ion monitoring: J Chromatogr B Analyt Technol Biomed Life Sci, 2018; 1085; 30-35
8. Garcia MCF, Soares DC, Santana RC: Parasitology, 2018; 145(9); 1219-27
9. Liu T, Wang CJ, Xie HQ, Guaiol – a naturally occurring insecticidal sesquiterpene: Nat Prod Commun, 2013; 8(10); 1353-54
10. An NTG, Huong LT, Satyal P: Plants (Basel), 2020; 9(4); 544
11. Garcia MCF, Soares DC, Santana RC: Parasitology, 2018; 145(9); 1219-27
12. Choudhary MI, Batool I, Atif M, Microbial transformation of (−)-guaiol and antibacterial activity of its transformed products: J Nat Prod, 2007; 70(5); 849-52
13. Soltani S, Amin G, Salehi-Sourmaghi MH, Histone deacetylase inhibitory and cytotoxic activities of the constituents from the roots of three species of Ferula: Iran J Basic Med Sci, 2019; 22(1); 93-98
14. Park HW, Choi SU, Baek NI: Arch Pharm Res, 2006; 29; 131-34
15. Yang Q, Wu J, Luo Y, (−)-Guaiol regulates RAD51 stability via autophagy to induce cell apoptosis in non-small cell lung cancer: Oncotarget, 2016; 7(38); 62585-97
16. Yang X, Zhu J, Wu J, (−)-Guaiol regulates autophagic cell death depending on mTOR signaling in NSCLC: Cancer Biol Ther, 2018; 19; 706-14
17. Zhang Y, Bao C, Mu Q, Reversal of cisplatin resistance by inhibiting PI3K/Akt signal pathway in human lung cancer cells: Neoplasma, 2016; 63(3); 362-70
18. Jia X, Wen Z, Sun Q, Apatinib suppresses the proliferation and apoptosis of gastric cancer cells via the PI3K/Akt signaling pathway: J BUON, 2019; 24(5); 1985-91
19. Ritchie ME, Phipson B, Wu D, limma powers differential expression analyses for RNA-sequencing and microarray studies: Nucleic Acids Res, 2015; 43(7); e47
20. Langfelder P, Horvath S, WGCNA: An R package for weighted correlation network analysis: BMC Bioinformatics, 2008; 9; 559
21. Wang Y, Xiao J, Suzek TO, PubChem’s BioAssay Database: Nucleic acids res, 2012; 40(Database issue); D400-12
22. Gong J, Cai C, Liu X, ChemMapper: A versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method: Bioinformatics, 2013; 29(14); 1827-29
23. Morgat A, Lombardot T, Coudert E, Enzyme annotation in UniProtKB using Rhea: Bioinformatics, 2020; 36(6); 1896-901
24. Chen H, Boutros PC, VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R: BMC Bioinformatics, 2011; 12; 35
25. Martin A, Ochagavia ME, Rabasa LC, BisoGenet: A new tool for gene network building, visualization and analysis: BMC Bioinformatics, 2010; 11; 91
26. Liu X, Liu X, Qiao T, Identification of crucial genes and pathways associated with colorectal cancer by bioinformatics analysis: Oncol Lett, 2020; 19(3); 1881-89
27. Primmer C, Papakostas S, Leder E, Annotated genes and nonannotated genomes: Cross-species use of Gene Ontology in ecology and evolution research, 2013; 22(12); 3216-41
28. Kanehisa M, Furumichi M, Tanabe M, KEGG: New perspectives on genomes, pathways, diseases and drugs, 2017; 45; D353-61
29. Yu G, Wang LG, Han Y, clusterProfiler: An R package for comparing biological themes among gene clusters: OMICS, 2012; 16(5); 284-87
30. Pinzi L, Rastelli G, Molecular docking: Shifting paradigms in drug discovery: Int J Mol Sci, 2019; 20(18); 4331
31. Burley SK, Berman HM, Kleywegt GJ, Protein Data Bank (PDB): The single global macromolecular structure archive: Methods Mol Biol, 2017; 1607; 627-41
32. Ito K, Murphy D, Application of ggplot2 to pharmacometric graphics: CPT Pharmacometrics Syst Pharmacol, 2013; 2(10); e79
33. Lortet-Tieulent J, Soerjomataram I, Ferlay J, International trends in lung cancer incidence by histological subtype: Adenocarcinoma stabilizing in men but still increasing in women: Lung Cancer, 2014; 84(1); 13-22
34. Akalin PK, Introduction to bioinformatics: Mol Nutr Food Res, 2006; 50(7); 610-19
35. Huang H, Regan KM, Lou Z, CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage: Science, 2006; 314(5797); 294-97
36. Kawakami M, Mustachio LM, Rodriguez-Canales J, Next-generation CDK2/9 inhibitors and anaphase catastrophe in lung cancer: J Natl Cancer Inst, 2017; 109(6); djw297
37. Denisenko TV, Budkevich IN, Zhivotovsky B, Cell death-based treatment of lung adenocarcinoma: Cell Death Dis, 2018; 9; 117
38. Liu X, Wang P, Zhang C, Epidermal growth factor receptor (EGFR): A rising star in the era of precision medicine of lung cancer: Oncotarget, 2017; 8(30); 50209-20
39. Li L, Zhu Z, Zhao Y, FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics: Sci Rep, 2019; 9(1); 7827
40. Dong Z, Yang P, Qiu X, KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis: J Cell Physiol, 2019; 234; 11304-14
41. Wang M, Feng L, Li PHsp90AB1 protein is overexpressed in non-small cell lung cancer tissues and associated with poor prognosis in lung adenocarcinoma patients: Zhongguo Fei Ai Za Zhi, 2016; 19; 64-69 [in Chinese]
42. Noorolyai SN, Shajari E, Baghbani , The relation between PI3K/AKT signalling pathway and cancer: Gene, 2019; 698; 120-28
43. Tan AC, Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC): Thorac Cancer, 2020; 11; 511-18
44. Engelman JA, Targeting PI3K signalling in cancer: Opportunities, challenges and limitations: Nat Rev Cancer, 2009; 9; 550-62
45. Scheffler M, Bos M, Gardizi M, PIK3CA mutations in non-small cell lung cancer (NSCLC): Genetic heterogeneity, prognostic impact and incidence of prior malignancies: Oncotarget, 2015; 6; 1315-26
46. Pérez-Ramírez C, Cañadas-Garre M, Molina MÁ, PTEN and PI3K/AKT in non-small-cell lung cancer: Pharmacogenomics, 2015; 16(16); 1843-62
47. Yamamoto H, Shigematsu H, Nomura M, PIK3CAmutations and copy number gains in human lung cancers: Cancer Res, 2008; 68; 6913-21
48. Tsujimoto Y, Role of Bcl-2 family proteins in apoptosis: Apoptosomes or mitochondria?: Genes Cells, 1998; 3(11); 697-707
49. Ashkenazi A, Fairbrother WJ, Leverson JD, From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors: Nat Rev Drug Discov, 2017; 16(4); 273-84
Figures
 Figure 1. In the flow chart, the systems pharmacology approach was used to determine the mechanism of (−)-Guaiol in the treatment of LUAD by integrating target identification, network construction, enrichment analysis, molecular docking and experimental validations.
Figure 1. In the flow chart, the systems pharmacology approach was used to determine the mechanism of (−)-Guaiol in the treatment of LUAD by integrating target identification, network construction, enrichment analysis, molecular docking and experimental validations. Figure 2. (A) Volcano plot of DEGs in TCGA. (B) Heat map of DEGs in TCGA. The green parts present down-regulated genes, red parts present up-regulated genes, and black parts presents non-differential genes. (C) The cluster dendrogram with different colors of TCGA. (D) Module-trait relationships. Each row corresponds to a color, while each column corresponds to the normal or tumor group.
Figure 2. (A) Volcano plot of DEGs in TCGA. (B) Heat map of DEGs in TCGA. The green parts present down-regulated genes, red parts present up-regulated genes, and black parts presents non-differential genes. (C) The cluster dendrogram with different colors of TCGA. (D) Module-trait relationships. Each row corresponds to a color, while each column corresponds to the normal or tumor group. Figure 3. Disease-drug-target network. Purple triangular node presents the drug, orange rhombic node presents disease, and green oval nodes present 480 targets.
Figure 3. Disease-drug-target network. Purple triangular node presents the drug, orange rhombic node presents disease, and green oval nodes present 480 targets. Figure 4. (A) Venn diagram was used to summarize the co-expression genes of DEGs, WGCNA module genes, and (−)-Guaiol target genes. (B) The 23 hub genes were predicted by topological screening of the protein–protein interaction (PPI) network with screening 2 times. DC – degree; BC – betweenness.
Figure 4. (A) Venn diagram was used to summarize the co-expression genes of DEGs, WGCNA module genes, and (−)-Guaiol target genes. (B) The 23 hub genes were predicted by topological screening of the protein–protein interaction (PPI) network with screening 2 times. DC – degree; BC – betweenness. Figure 5. (A) Representative results (P<0.05) of GO enrichment analysis. (B) Representative results (p<0.05) of KEGG enrichment analyses. Bubble size indicates gene count and the color indicates P value.
Figure 5. (A) Representative results (P<0.05) of GO enrichment analysis. (B) Representative results (p<0.05) of KEGG enrichment analyses. Bubble size indicates gene count and the color indicates P value. Figure 6. KEGG pathway-target network. The orange round nodes represent the top 19 related targets and the green round nodes represent the top 20 pathways.
Figure 6. KEGG pathway-target network. The orange round nodes represent the top 19 related targets and the green round nodes represent the top 20 pathways. Figure 7. Molecular docking diagrams of (−)-Guaiol with (A) CDK2, (B) EGFR, (C) HSP90AA1, (D) HSP90AB1, and (E) FN1. (F) The binding affinity for the 5 targets docked into the (−)-Guaiol crystal structure.
Figure 7. Molecular docking diagrams of (−)-Guaiol with (A) CDK2, (B) EGFR, (C) HSP90AA1, (D) HSP90AB1, and (E) FN1. (F) The binding affinity for the 5 targets docked into the (−)-Guaiol crystal structure. Figure 8. (A) Cell survival rate was detected by performing a CCK-8 assay. The LUAD cells (A549 and Calu-1) and normal lung cells BEAS-2B cells treated with different concentrations of (−)-Guaiol. IC50 values were 75.7 μM for A549 cells, 92.1 μM for H1299 cells, and 322.3 μM for BEAS-2B cells. Data from 3 independent experiments were represented as means±SD. (B) A549 and Calu-1 were treated with (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) companied Z-VAD (20μM) for 24 h. ** P<0.01 vs control in A549. # P<0.05 vs (−)-Guaiol in Calu-1. Data from 3 independent experiments were represented as means±SD. (C) A549 and Calu-1 were treated with or without (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) and Z-VAD (20 μM), and stained with SYTOX Green (100 nM), then observed under a fluorescence microscope.
Figure 8. (A) Cell survival rate was detected by performing a CCK-8 assay. The LUAD cells (A549 and Calu-1) and normal lung cells BEAS-2B cells treated with different concentrations of (−)-Guaiol. IC50 values were 75.7 μM for A549 cells, 92.1 μM for H1299 cells, and 322.3 μM for BEAS-2B cells. Data from 3 independent experiments were represented as means±SD. (B) A549 and Calu-1 were treated with (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) companied Z-VAD (20μM) for 24 h. ** P<0.01 vs control in A549. # P<0.05 vs (−)-Guaiol in Calu-1. Data from 3 independent experiments were represented as means±SD. (C) A549 and Calu-1 were treated with or without (−)-Guaiol (75 μM) or (−)-Guaiol (75 μM) and Z-VAD (20 μM), and stained with SYTOX Green (100 nM), then observed under a fluorescence microscope. Figure 9. (A) Flow cytometry to detect the apoptosis rate in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (B) Fluorescence microscope was used to observe apoptosis in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (C) Western blot analysis was used to detect the protein expression of BAX and Bcl-2 in A549 treated with (−)-Guaiol (60 μM or 120 μM). The relative BAX and Bcl-2 levels from 3 independent experiments were statistically analyzed using ANOV A test in the quantitative analysis. * P<0.05, *** P<0.001 vs control.
Figure 9. (A) Flow cytometry to detect the apoptosis rate in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (B) Fluorescence microscope was used to observe apoptosis in A549 and Calu-1 treated with different concentrations of (−)-Guaiol. (C) Western blot analysis was used to detect the protein expression of BAX and Bcl-2 in A549 treated with (−)-Guaiol (60 μM or 120 μM). The relative BAX and Bcl-2 levels from 3 independent experiments were statistically analyzed using ANOV A test in the quantitative analysis. * P<0.05, *** P<0.001 vs control. Figure 10. (−)-Guaiol regulates the expression of related proteins in PI3K/Akt Signaling Pathway. Western blot analysis was used to detect the protein expression of PI3K, p-PI3K, Akt, and p-Akt in A549 treated with or without (−)-Guaiol (75 μM) or LY294002 (15 μM). The relative protein levels from 3 independent experiments were statistically analyzed using ANOVA in the quantitative analysis. * P<0.05, ** P<0.01, *** P<0.001 vs control. ## P<0.01, ### P<0.001 vs LY294002.
Figure 10. (−)-Guaiol regulates the expression of related proteins in PI3K/Akt Signaling Pathway. Western blot analysis was used to detect the protein expression of PI3K, p-PI3K, Akt, and p-Akt in A549 treated with or without (−)-Guaiol (75 μM) or LY294002 (15 μM). The relative protein levels from 3 independent experiments were statistically analyzed using ANOVA in the quantitative analysis. * P<0.05, ** P<0.01, *** P<0.001 vs control. ## P<0.01, ### P<0.001 vs LY294002. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952