27 April 2022: Clinical Research
Differences Between Sorafenib and Lenvatinib Treatment from Genetic and Clinical Perspectives for Patients with Hepatocellular Carcinoma
Lei Wang1CD, Lei Wang23DEF, Bo Xiao4BC, Mingxuan Cui4CDE, Bo Zhang4A*DOI: 10.12659/MSM.934936
Med Sci Monit 2022; 28:e934936
Abstract
BACKGROUND: The aim of this work was to systematically compare the differences between sorafenib and lenvatinib for patients with hepatocellular carcinoma (HCC) from genetic and clinical perspectives.
MATERIAL AND METHODS: The mRNA and miRNA sequencing information of patients with HCC treated with either sorafenib or lenvatinib was analyzed using differential expression and a protein–protein interaction assay. The clinical manifestations and adverse events of the 2 drugs were also investigated.
RESULTS: Compared with patients with HCC treated with sorafenib, patients treated with lenvatinib developed 8 differentially expressed genes (DEGs, FGF4, FGF23, UNC13C, RIMBP2, STXBP5L, PHOX2B, NEUROD4, and POU4F2) and 3 miRNAs (DEMs, has-miR-548ah, has-miR-888, and has-miR-196a-1), of which hsa-miR-548 regulated 4 target genes, the largest number among the 3 miRNAs. The functions of these DEMs and DEGs were verified by external experiments in the HCC cell line Hep3B2.1-7. We further investigated the adverse events of the drugs for patients with advanced HCC in clinical treatment. The patients in the sorafenib group developed less frequent symptoms of hypertension and diarrhea. Also, the frequency of hand-foot skin reactions in patients treated with lenvatinib was lower than that of patients treated with sorafenib (P<0.05). There were no significant differences in nausea, fatigue, frequent urination, and dizziness (P>0.05).
CONCLUSIONS: In a time of increasing interest in chemotherapy drug treatments for patients with HCC, this study provided a better understanding of the clinical evaluations of sorafenib and lenvatinib.
Keywords: Carcinoma, Hepatocellular, lenvatinib, sorafenib, Antineoplastic Agents, Humans, Liver Neoplasms, MicroRNAs, Phenylurea Compounds, Quinolines
Background
Hepatocellular carcinoma (HCC) is the most frequent primary malignancy of the liver, comprising 75% to 85% of cases of liver cancer [1], creating a critical medical problem worldwide. There were 782 000 cases diagnosed and 746 000 deaths in 2012, giving HCC the rank as the sixth most common neoplasm and the second leading cause of cancer-related deaths [2]. The worldwide incidence of HCC is heterogeneous owing to the prevalence of risk factors. The formation of HCC is closely associated with the presence of chronic liver disease [3]. The hepatitis B virus (HBV) and hepatitis C virus (HCV) are the primary causes, which subsequently lead to chronic liver disease [4]. The pathogenesis of virus-induced HCC has been suggested to be associated with various mechanisms, including the integration of HBV-DNA into the host’s genetic machinery, selective immunosuppression for virus presentation, down regulation of viral protection gene expression, virus-specific T-cell suppression for recognizing HBV antigens, and DNA methylation [5]. In addition to virus-induced HCC, growing evidence from retrospective studies supports the connection between HCC and other non-viral risk factors, such as diabetes, alcoholism, and dyslipidemia, especially in developed regions [6]. For most patients, the disorder is diagnosed at an advanced stage, when surgical treatment is no longer an option. Clinically, patients with advanced HCC need chemotherapy to improve treatment. Based on decades of efforts, researchers have provided several potential systemic therapies targeting advanced HCC, namely sorafenib, lenvatinib, regorafenib, cabozantinib, atezolizumab plus bevacizumab, and ramucirumab in phase III trials [7].
Until 2008, there was still no effective therapy for patients diagnosed with advanced-stage HCC or patients who transitioned into it as other therapies failed. Sorafenib, developed by the Bayer and Onyx companies, was initially approved by the FDA for advanced HCC treatment in 2006 [8]. A year later, it proved to be a unique target drug for HCC [9,10]. Sorafenib is a small polytyrosine kinase inhibitor, which dominantly suppresses Raf kinase, vascular endothelial growth factor, and platelet-derived growth factor function [11]. Sorafenib was the first systemic therapy approved in HCC as the result of 2 positive randomized placebo-controlled trials, with 1 multicenter trial done predominantly in Europe and the United States, and the other trial done in the Asia-Pacific area. Lenvatinib is a receptor tyrosine kinase oral small-molecule inhibitor, which was recently approved for first-line treatment in patients with unresectable advanced HCC in the United States, the European Union, Japan, and China [12]. Lenvatinib was clinically initiated as a substitution for sorafenib. However, the molecular mechanism of lenvatinib is poorly understood. Meanwhile, it is still controversial whether lenvatinib could replace sorafenib since both of them cause diverse adverse events, including hand-foot skin reactions, arterial hypertension, fatigue, and diarrhea [13]. Currently, immune-based combinations of drugs (mainly targeting the immune checkpoint PD L-1) seem to have shifted the direction of future first-line therapies [14]. For instance, the combination of lenvatinib and pembrolizumab is now being evaluated as a front-line treatment in patients with advanced HCC, and the early phase clinical trials have already reported promising results [15,16].
Although new types of drugs have been discovered gradually, sorafenib and lenvatinib are still the mainstream therapies for patients with advanced HCC. However, the overall effects of sorafenib and lenvatinib are far from satisfactory, and the clinical therapy selection is still an issue owing to the unknown molecular mechanisms. To address these issues, in this study, a differential expression analysis was performed on patients with HCC treated with either sorafenib or lenvatinib. The profile of target mRNAs and associated miRNAs was also developed. The clinical symptoms and adverse reactions in patients with HCC for the 2 different drug administrations were further explored. This study provides potential guidance for the precise administration of sorafenib and lenvatinib in HCC treatment.
Material and Methods
PARTICIPANTS:
This study was a follow-up analysis of our previous study, which was given ethics approval, and all patients gave informed consent to participate [17]. All analyses of human data were carried out in strict compliance with relevant ethics regulations. Overall, 120 patients with advanced HCC who were admitted to our hospital from September 2019 to December 2020 were randomly selected to participant in the study. The medical records of all the recruited individuals were retrospectively reviewed. All individuals in this study were randomly selected based on the following inclusion criteria: (1) age from 40 to 80 years; (2) for laboratory measurement, the expression level of alpha-fetoprotein was greater than 400 μg/L over 1 month or 200 to 400 μg/L lasting for over 2 months; and (3) for imaging examination, the liver mass displayed more than 2 cm in diameter and typical HCC characteristics based on either computed tomography (CT) or magnetic resonance imaging (MRI) analysis. If the liver mass was 1 to 2 cm in diameter, CT and MRI were required. Participants were excluded if they had chronic liver disease or the presence of a malignancy other than HCC. The recruited individuals were untreated with chemotherapy for the previous 3 months.
The treatment rationale was as follows: 70 patients were treated with sorafenib, 40 patients were treated with lenvatinib, and 10 patients had no chemotherapy drug treatment, starting at the same time point. There was no significant difference in basic clinical data between these groups. For sorafenib administration, hospitalized patients with advanced HCC were treated with sorafenib tosylate tablets (Bayer, Germany) at a dosage of 0.4 g twice daily on an empty stomach or with a low-fat or medium-fat diet for 6 months. At the same time, for lenvatinib administration, hospitalized patients with advanced HCC were treated with lenvatinib mesylate capsules (Merck & Co, Inc, USA) at a dosage as 12 mg once daily on an empty stomach or with food for 6 months.
DIFFERENTIALLY EXPRESSED MRNA AND MIRNA ANALYSIS:
According to a previous report [18], the cancerous tissues of all patients with HCC were dissected and snap-frozen during surgery after the chemotherapy treatment and stored in liquid nitrogen immediately. The total RNA was extracted using an RNAiso Plus kit (Takara, Beijing, China). The total RNA samples were collected and the mRNA and miRNA expression were analyzed by Geneseed Co (Guangdong, China). The expression analysis was conducted using the Affymetrix GeneChip Pico kit and hybridized to Affymetrix Clariom S arrays as described by the manufacturer (Affymetrix, USA).
The robust multi-array average method was first utilized to normalize the original data measured by the chip. After that, the normalized value was calculated using the log2 logarithm to provide the data after normalization, following differential expression analysis. The edgeR package in R language was developed to analyze differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) between different treatments. The absolute values of logarithmic transformed differential expression multiples (Log2FC) (used for DEGs and DEMs screening) were the indications of absolute values for different selected targets. The values of Log2FC >1 and P<0.05 were used as criteria for screening [19]. The P value in the results represented the adjusted P values (false discovery rate) with considering multiple comparison criterion, which was between all the genes and miRNA together in expression analysis.
PROTEIN–PROTEIN INTERACTION NETWORKS AND ANALYSIS OF MIRNA TARGET GENES:
The STRING (https://string-db.org/, version 11.0) was generated to analyze the functional connections and interactions of candidate proteins [20]. Cytoscape (https://cytoscape.org/, version 3.7.2) analysis was performed to visualize the protein–protein interaction (PPI) network [21].
The target genes of miRNAs were predicted through the miRDB (http://mirdb.org/index.html, version 6.0) database [22]. Meanwhile, Cytoscape was used to visualize the miRNA-mRNA regulatory network.
CELL CULTURE:
The human HCC cell line Hep3B2.1–7 was purchased from American Type Culture Collection Co (China). The cells were cultured at 37 °C in 5% CO2 in Eagle’s minimum essential medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin, based on the manufacturer’s instruction. As previously reported [23], the cells were treated with either sorafenib or lenvatinib at 3 μM for 4 days.
For functional experiments, a specific has-miR-548ah mimic/inhibitor (Ambion) (50 nmol/L) and a corresponding negative control (Ambion) were transfected into the Hep3B2.1–7 cells. The transfection was achieved using Lipofectamine 2000 (Sigma-Aldrich, Beijing, China).
QUANTITATIVE REAL-TIME POLYMERASE CHAIN REACTION:
The total RNA from the cancerous tissues of patients with HCC were extracted using an RNAiso Plus kit (Takara, Beijing, China). The TRIzol reagent was used to extract total RNA from the HCC cell line Hep3B2.1–7. The diverse expression activities of potential target genes were compared using the quantitative real-time polymerase chain reaction (qPCR) method. The high-capacity RNA to cDNA kit (Applied Biosytems) was used to reversely transcribe RNA (1000 ng) to cDNA, and qPCR amplification was performed with SYBR Green (Qiagen). The resulting value was normalized according to the internal reference gene GAPDH of each parallel sample. For miRNA expression analysis, the TaqMan advanced MicroRNA assay kit (Applied Biosystems) and miRNA-specific primers reverse transcription RNA (100 ng) were used in this study. PCR conditions were set up according to the instructions: 1 cycle at 95°C 5 min; 40 cycles at 95°C for 15 s; followed by 60°C for 40 s. The relative expression was measured based on the 2-ΔΔCt method. All experiments were performed in triplicate. The DEGs primers were designed based on the PrimerBank site (
The final PCR conditions were set up as follows: 1 cycle at 95°C 5 min; 40 cycles at 95°C for 15 s; followed by 60°C for 40 s. The RT-PCR relative expression analysis was conducted using the 2-ΔΔCt method with GAPDH as an internal control gene. All experiments were performed 3 times independently.
STATISTICAL ANALYSIS:
SAS 9.4 and SPSS 19.0 statistical software were used for data analysis. The continuous variables were tested for normal distribution, and the variables conforming to the normal distribution were expressed as mean±standard deviation. The
Results
COMPARISONS OF DEGS AND DEMS FOR THE 2 DRUGS:
Following the previously reported procedure [18], RNA samples from 10 patients treated with either sorafenib (sorafenib group, 10 patients) or lenvatinib (lenvatinib group, 10 patients), and 10 control patients without any chemotherapy drug treatment (control group, 10 patients) were extracted and analyzed. At the same time, the edgeR package in R language was performed to analyze the DEGs and DEMs between the 2 different treatments. Firstly, we compared the expression levels of all mRNAs of the patients with HCC treated with sorafenib alone and patients with HCC without any chemotherapy. Simultaneously, we compared the expression levels of all mRNAs in patients treated with lenvatinib alone with those without any chemotherapy drugs. The sorafenib-treated patients displayed 466 DEGs, including 57 upregulated genes and 409 downregulated genes, compared with control group (Figure 1A). Meanwhile, lenvatinib-treated patients displayed 471 DEGs, including 65 upregulated genes and 406 downregulated genes (Figure 1B). By taking the intersection between sorafenib and lenvatinib, 8 genes were selected as the primary significant DEGs for lenvatinib: FGF4, FGF23, UNC13C, RIMBP2, STXBP5L, PHOX2B, NEUROD4, and POU4F2 (Figure 1E). FGF4 and FGF23 were primary DEGs for 95% of the lenvatinib-treated patients with HCC.
The miRNA expression was analyzed using the same method. Compared with the control group, patients treated with sorafenib alone displayed 12 DEMs, including 6 upregulated miRNAs and 6 downregulated miRNAs (Figure 1C); while the lenvatinib-treated group showed 12 differentially expressed DEMs, including 7 upregulated miRNAs and 5 downregulated miRNAs (Figure 1D). The miRNAs has-miR-548ah, has-miR-888, and has-miR-196a-1 were specific to lenvatinib and were all upregulated (Figure 1F). The miRNA has-miR-548ah was the primary DEM for 90% of the lenvatinib-treated patients with HCC.
ESTABLISHMENT OF MIRNA-MRNA REGULATORY NETWORK:
We utilized the STRING database to construct a PPI network for the 8 genes. The MCODE plug-ins were used to identify significant clustering modules for the gene interactions (Figure 2A). At the same time, the 3 candidate miRNAs and 8 mRNAs were further investigated for a regulatory network and visualized with Cytoscape software. Of the miRNAs, hsa-miR-548 regulated the largest number of target genes, at 4. Meanwhile, the number of target genes regulated by hsa-miR-888 and hsa-miR-196a-1 were both 2 (Figure 2B). Based on these facts, the 3 DEMs and 8 DEGs were suggested to be the potential hub genes for lenvatinib treatment in patients with HCC.
EXTERNAL IN VITRO EXPERIMENTS VERIFICATION OF POTENTIAL DEGS AND DEMS FOR LENVATINIB:
Next, we sought to verify the functions of potential DEGs and DEMs for lenvatinib based on in vitro analysis. The HCC cell line Hep3B2.1–7 was treated with either sorafenib or lenvatinib at 3 μM for 4 days. Compared with that of the untreated control, the expression levels of DEGs (FGF4, FGF23, UNC13C, RIMBP2, STXBP5L, PHOX2B, NEUROD4, and POU4F2) were decreased based on the RT-PCR measurement (Figure 2C); while the activities of DEMs (has-miR-548ah, has-miR-888, and has-miR-196a-1) were enhanced (Figure 2D). At the same time, sorafenib treatment did not show significant differences for the DEGs and DEMs of lenvatinib (data not shown). These results were consistent with the results of the differential expression analysis (Figure 1).
Moreover, given the expression level of FGF4 as a read-out, the mimic transfection of has-miR-548ah in Hep3B2.1–7 cells significantly reduced the activity of FGF4. Reversely, has-miR-548ah inhibitor transfection elevated the activity of FGF4. Meanwhile, the has-miR-548ah inhibitor attenuated the function of lenvatinib for the expression level of FGF4 (Figure 2E). Collectively, these outcomes suggested that has-miR-548ah was a central downstream factor of lenvatinib for FGF4 gene regulation.
COMPARISON OF CLINICAL TUMOR MARKERS AND HEPATOBILIARY FUNCTION BETWEEN THE 2 DRUGS:
In addition to the molecular mechanism network, we further compared the clinical manifestations of the 2 treatments for advanced patients with HCC. There was no obvious difference in the first month between sorafenib and lenvatinib treatment. However, there were significant differences in alpha-fetoprotein, carbohydrate antigen 19-9, alanine aminotransferase, aspartate aminotransferase, total bilirubin, and direct bilirubin among patients in the lenvatinib group (P<0.05) from the third month, which suggested that lenvatinib had a better therapeutic effect than sorafenib (Table 1). During the observation period, the cholinesterase levels of the sorafenib and lenvatinib groups remained similar (P>0.05).
COMPARISON OF CLINICAL ADVERSE REACTIONS BETWEEN THE 2 DRUGS:
Adverse effects of both drugs occurred frequently. To address the advantages and disadvantages of the 2 treatments in terms of adverse effects, we further investigated the adverse events associated with the treatments, including hand-foot skin reaction, arterial hypertension, diarrhea, fatigue, nausea, frequent urination, and dizziness, for patients with advanced HCC. As shown in Table 2, no serious adverse events occurred for the patients in the 2 groups. However, patients in the sorafenib group developed hypertension and diarrhea less frequently than the lenvatinib group. At the same time, the frequency of hand-foot skin reaction in patients treated with lenvatinib was relatively lower (P<0.05). There were no significant differences in nausea, fatigue, frequent urination, and dizziness in this study (P>0.05).
Discussion
LIMITATIONS:
This study was based on the medical records from a single center and the sample size was limited, which could affect the accuracy of the results. Also, we did not recruit patients with immune-based combinations of drugs for treatment of advanced HCC because the number of patients is small in our hospital and therefore, we could not investigate the differences between single-drug and combined-drug therapy. Moreover, we could not provide a conclusion for long-term clinical manifestations of lenvatinib vs sorafenib for patients with advanced HCC after 3 months of continuous drug treatment. These issues may be worth exploring in the next study.
Conclusions
In this study, we systematically compared the differences between sorafenib and lenvatinib for patients with advanced HCC from genetic and clinical perspectives. Patients treated with lenvatinib developed 8 DEGs (
Figures
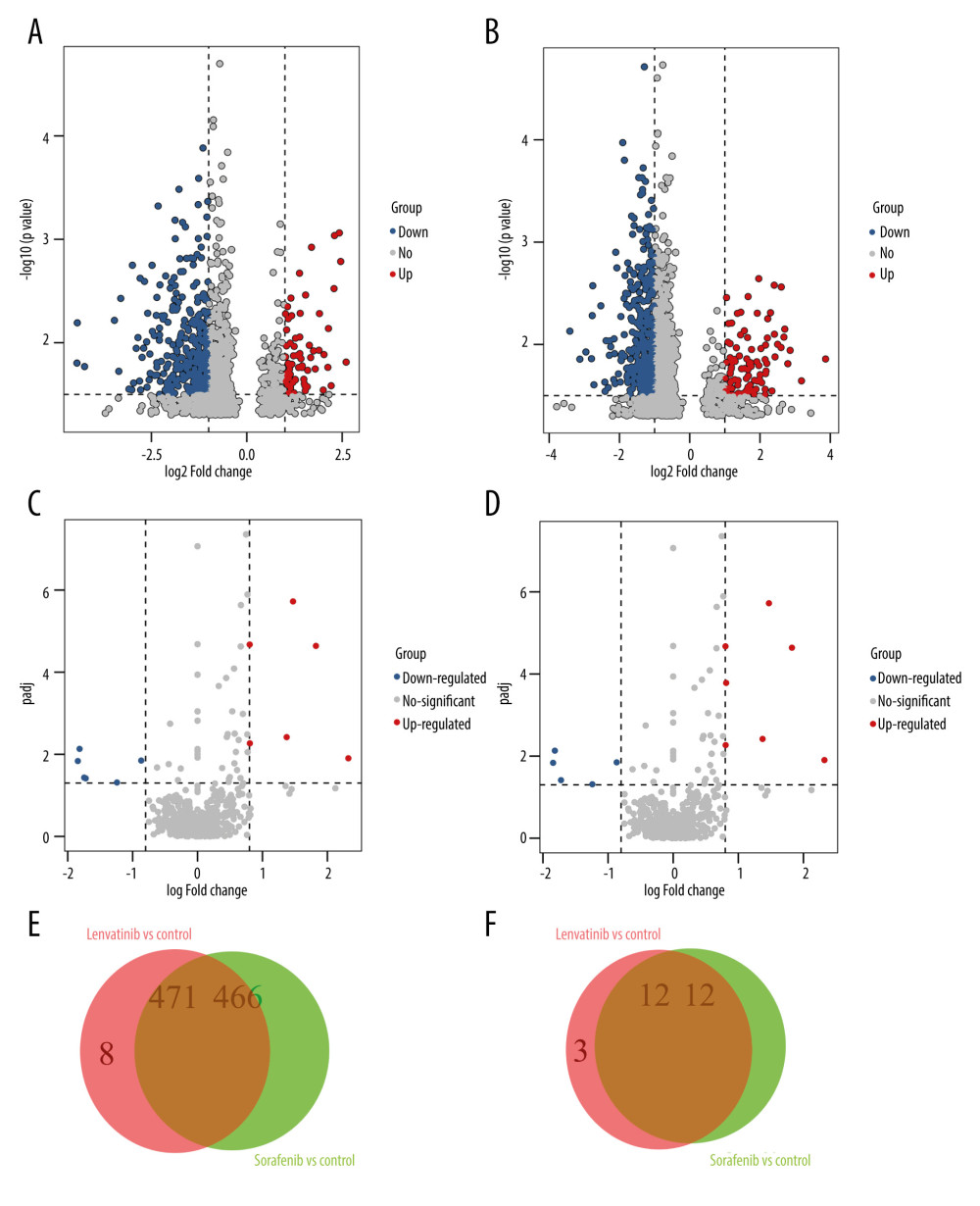 Figure 1. The differential analysis of mRNA and miRNA. (A, B) Volcano diagram of differentially expressed genes (DEGs) for Sorafenib treatment and Lenvatinib treatment, respectively. The horizontal axis is the log2FC value, while the vertical axis is −log10 (P value). Red color represents upregulation, green represents downregulation, and black represents no significant difference respectively. (C, D) The volcano diagram of differentially expressed miRNAs (DEMs). (E, F) The Venn diagrams of DEGs and DEMs for Sorafenib treatment and Lenvatinib treatment, respectively.
Figure 1. The differential analysis of mRNA and miRNA. (A, B) Volcano diagram of differentially expressed genes (DEGs) for Sorafenib treatment and Lenvatinib treatment, respectively. The horizontal axis is the log2FC value, while the vertical axis is −log10 (P value). Red color represents upregulation, green represents downregulation, and black represents no significant difference respectively. (C, D) The volcano diagram of differentially expressed miRNAs (DEMs). (E, F) The Venn diagrams of DEGs and DEMs for Sorafenib treatment and Lenvatinib treatment, respectively. 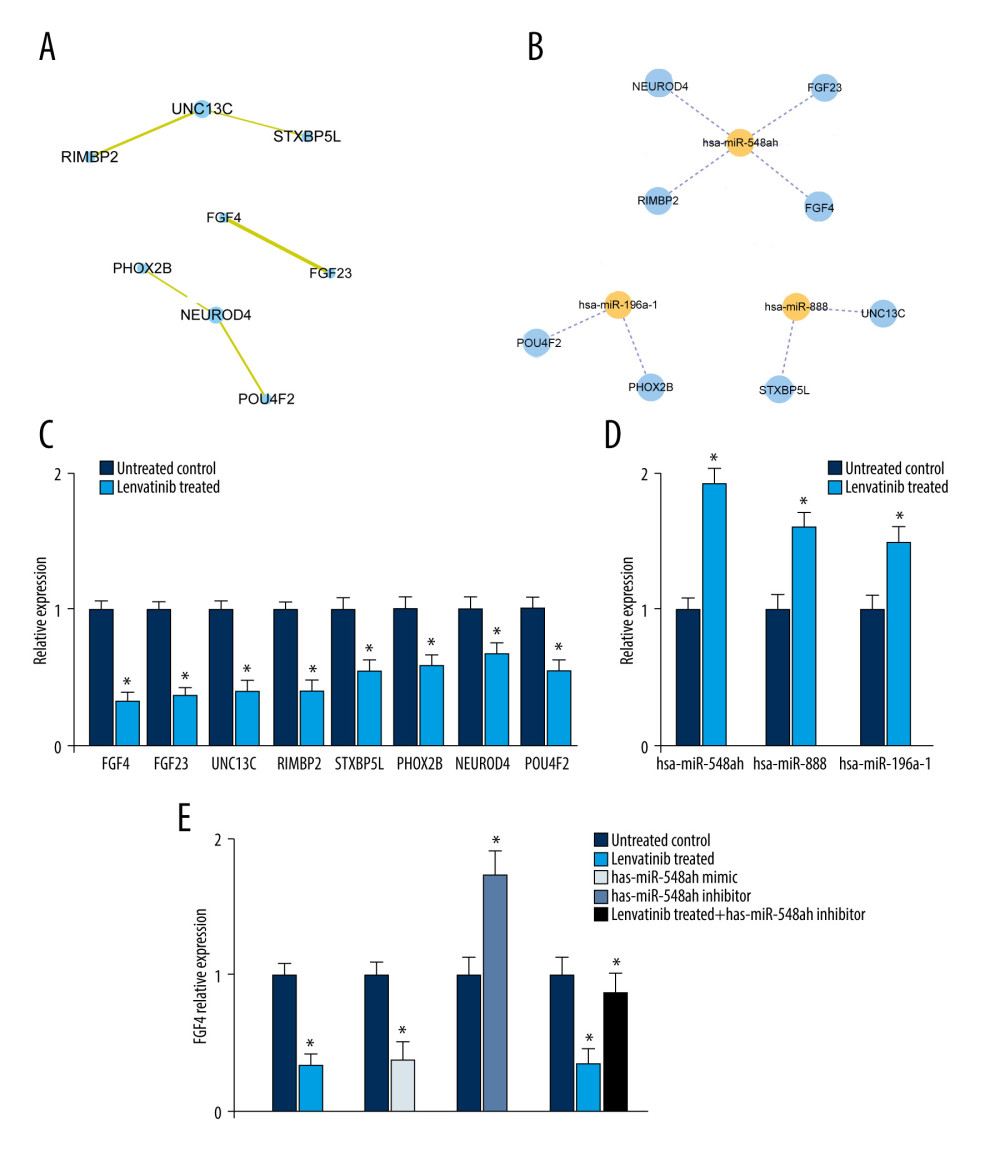 Figure 2. Establishment of protein–protein interaction (PPI) and miRNA-mRNA regulatory network. (A) The construction of PPI network, in which each dot represents a node. (B) The miRNA-mRNA regulatory network. The orange node is miRNA and the blue node is mRNA. The solid line represents positive regulation, and the dotted line represents negative regulation respectively. (C) The RT-PCR investigations of differentially expressed genes. (D) The RT-PCR investigations of differentially expressed miRNAs. (E) The expression levels of FGF4 for different treatments in Hep3B2.1–7 cells.
Figure 2. Establishment of protein–protein interaction (PPI) and miRNA-mRNA regulatory network. (A) The construction of PPI network, in which each dot represents a node. (B) The miRNA-mRNA regulatory network. The orange node is miRNA and the blue node is mRNA. The solid line represents positive regulation, and the dotted line represents negative regulation respectively. (C) The RT-PCR investigations of differentially expressed genes. (D) The RT-PCR investigations of differentially expressed miRNAs. (E) The expression levels of FGF4 for different treatments in Hep3B2.1–7 cells. References
1. Colquhoun SD, Hepatocellular carcinoma: The current role of surgical intervention: Crit Rev Oncog, 2016; 21(1); 93-103
2. Forner A, Reig M, Bruix J, Hepatocellular carcinoma: Lancet, 2018; 391(10127); 1301-14
3. Kulik L, El-Serag HB, Epidemiology and management of hepatocellular carcinoma: Gastroenterology, 2019; 156(2); 477-91
4. Armengol C, Sarrias MR, Sala M, Hepatocellular carcinoma: Present and future: Med Clin (Barc), 2018; 150(10); 390-97
5. Ozer-Etik D, Suna N, Boyacioglu AS, Management of hepatocellular carcinoma: Prevention, surveillance, diagnosis, and staging: Exp Clin Transplant, 2017; 15(Suppl 2); 31-35
6. Golabi P, Rhea L, Henry L, Hepatocellular carcinoma and non-alcoholic fatty liver disease: Hepatol Int, 2019; 13; 688-94
7. Llovet JM, Kelley RK, Villanueva A, Hepatocellular carcinoma: Nat Rev Dis Primers, 2021; 7(1); 6
8. Rocca A, Sorafenib for the treatment of breast cancer: Expert Opin Pharmacother, 2017; 18(6); 621-30
9. Keating GM, Sorafenib: A review in hepatocellular carcinoma: Target Oncol, 2017; 12(2); 243-53
10. Mendez-Blanco C, Fondevila F, Garcia-Palomo A, Sorafenib resistance in hepatocarcinoma: role of hypoxia-inducible factors: Exp Mol Med, 2018; 50(10); 1-9
11. Zhu YJ, Zheng B, Wang HY, New knowledge of the mechanisms of sorafenib resistance in liver cancer: Acta Pharmacol Sin, 2017; 38(5); 614-22
12. Al-Salama ZT, Syed YY, Scott LJ, Lenvatinib: A review in hepatocellular carcinoma: Drugs, 2019; 79(6); 665-74
13. Suyama K, Iwase H, Lenvatinib: A promising molecular targeted agent for multiple cancers: Cancer Control, 2018; 25(1); 1073274818789361
14. Rizzo A, Ricci AD, Brandi G, Immune-based combinations for advanced hepatocellular carcinoma: Shaping the direction of first-line therapy: Future Oncol, 2021; 17(7); 755-57
15. Rizzo A, Dadduzio V, Ricci AD, Lenvatinib plus pembrolizumab: The next frontier for the treatment of hepatocellular carcinoma?: Expert Opin Investig Drugs, 2021; 30(1); 1-8
16. Rizzo A, Ricci AD, Brandi G, Atezolizumab in advanced hepatocellular carcinoma: Good things come to those who wait: Immunotherapy, 2021; 13(8); 637-44
17. Xu J, Shen X, Zhang B, Development and validation of LRP1B mutation-associated prognostic model for hepatocellular carcinoma: Biosci Rep, 2021; 41(9); BSR20211053
18. Hoshi T, Watanabe-Miyano S, Watanabe H, Lenvatinib induces death of human hepatocellular carcinoma cells harboring an activated FGF signaling pathway through inhibition of FGFR-MAPK cascades: Biochem Biophys Res Commun, 2019; 513(1); 1-7
19. Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data: Bioinformatics, 2010; 26(1); 139-40
20. Yu G, Wang LG, Han Y, clusterProfiler: An R package for comparing biological themes among gene clusters: Omics J Integr Biol, 2012; 16(1); 284-87
21. Szklarczyk D, Gable AL, Lyon D, STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47(1); D607-D13
22. Shannon P, Markiel A, Ozier O, Cytoscape: a software environment for integrated models of biomolecular interaction networks: Genome Res, 2003; 13(1); 2498-504
23. Mathew NR, Baumgartner F, Braun L, Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells: Nat Med, 2018; 24(3); 282-91
24. Hartke J, Johnson M, Ghabril M, The diagnosis and treatment of hepatocellular carcinoma: Semin Diagn Pathol, 2017; 34(2); 153-59
25. Khemlina G, Ikeda S, Kurzrock R, The biology of Hepatocellular carcinoma: Implications for genomic and immune therapies: Mol Cancer, 2017; 16(1); 149
26. Matsui J, Yamamoto Y, Funahashi Y, E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition: Int J Cancer, 2008; 122(3); 664-71
27. Cabanillas ME, Habra MA, Lenvatinib: Role in thyroid cancer and other solid tumors: Cancer Treat Rev, 2016; 42; 47-55
28. Hao Z, Wang P, Lenvatinib in management of solid tumors: Oncologist, 2020; 25(2); e302-310
29. Xing TJ, Xu HT, Yu WQ, MiRNA-548ah, a potential molecule associated with transition from immune tolerance to immune activation of chronic hepatitis B: Int J Mol Sci, 2014; 15(8); 14411-26
30. Li YB, Sun FN, Ma XY, MiR-888 promotes cell migration and invasion of hepatocellular carcinoma by targeting SMAD4: Eur Rev Med Pharmacol Sci, 2019; 23(5); 2020-27
31. Cao JX, miR888 regulates cancer progression by targeting multiple targets in lung adenocarcinoma: Oncol Rep, 2019; 41(6); 3367-76
32. Gao SJ, Chen L, Lu W, miR-888 functions as an oncogene and predicts poor prognosis in colorectal cancer: Oncol Lett, 2018; 15(6); 9101-9
33. Feng C, She J, Chen X, Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP1: Nanomedicine (Lond), 2019; 14(19); 2579-93
34. Katoh M, FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review): Int J Mol Med, 2016; 38(1); 3-15
35. Rezzola S, Ronca R, Loda A, The Autocrine FGF/FGFR System in both skin and uveal melanoma: FGF trapping as a possible therapeutic approach: Cancers (Basel), 2019; 11(9); 1305
36. Velmurugan BK, Yeh KT, Hsieh MJ, UNC13C suppress tumor progression via inhibiting EMT pathway and improves survival in oral squamous cell carcinoma: Front Oncol, 2019; 9; 728
37. Naftali O, Maman S, Meshel T, PHOX2B is a suppressor of neuroblastoma metastasis: Oncotarget, 2016; 7; 10627-37
38. Personeni N, Pressiani T, Rimassa L, Lenvatinib for the treatment of unresectable hepatocellular carcinoma: Evidence to date: J Hepatocell Carcinoma, 2019; 6; 31-39
39. Kudo M, Finn RS, Qin S, Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial: Lancet, 2018; 391(10126); 1163-73
Figures
 Figure 1. The differential analysis of mRNA and miRNA. (A, B) Volcano diagram of differentially expressed genes (DEGs) for Sorafenib treatment and Lenvatinib treatment, respectively. The horizontal axis is the log2FC value, while the vertical axis is −log10 (P value). Red color represents upregulation, green represents downregulation, and black represents no significant difference respectively. (C, D) The volcano diagram of differentially expressed miRNAs (DEMs). (E, F) The Venn diagrams of DEGs and DEMs for Sorafenib treatment and Lenvatinib treatment, respectively.
Figure 1. The differential analysis of mRNA and miRNA. (A, B) Volcano diagram of differentially expressed genes (DEGs) for Sorafenib treatment and Lenvatinib treatment, respectively. The horizontal axis is the log2FC value, while the vertical axis is −log10 (P value). Red color represents upregulation, green represents downregulation, and black represents no significant difference respectively. (C, D) The volcano diagram of differentially expressed miRNAs (DEMs). (E, F) The Venn diagrams of DEGs and DEMs for Sorafenib treatment and Lenvatinib treatment, respectively. Figure 2. Establishment of protein–protein interaction (PPI) and miRNA-mRNA regulatory network. (A) The construction of PPI network, in which each dot represents a node. (B) The miRNA-mRNA regulatory network. The orange node is miRNA and the blue node is mRNA. The solid line represents positive regulation, and the dotted line represents negative regulation respectively. (C) The RT-PCR investigations of differentially expressed genes. (D) The RT-PCR investigations of differentially expressed miRNAs. (E) The expression levels of FGF4 for different treatments in Hep3B2.1–7 cells.
Figure 2. Establishment of protein–protein interaction (PPI) and miRNA-mRNA regulatory network. (A) The construction of PPI network, in which each dot represents a node. (B) The miRNA-mRNA regulatory network. The orange node is miRNA and the blue node is mRNA. The solid line represents positive regulation, and the dotted line represents negative regulation respectively. (C) The RT-PCR investigations of differentially expressed genes. (D) The RT-PCR investigations of differentially expressed miRNAs. (E) The expression levels of FGF4 for different treatments in Hep3B2.1–7 cells. Tables
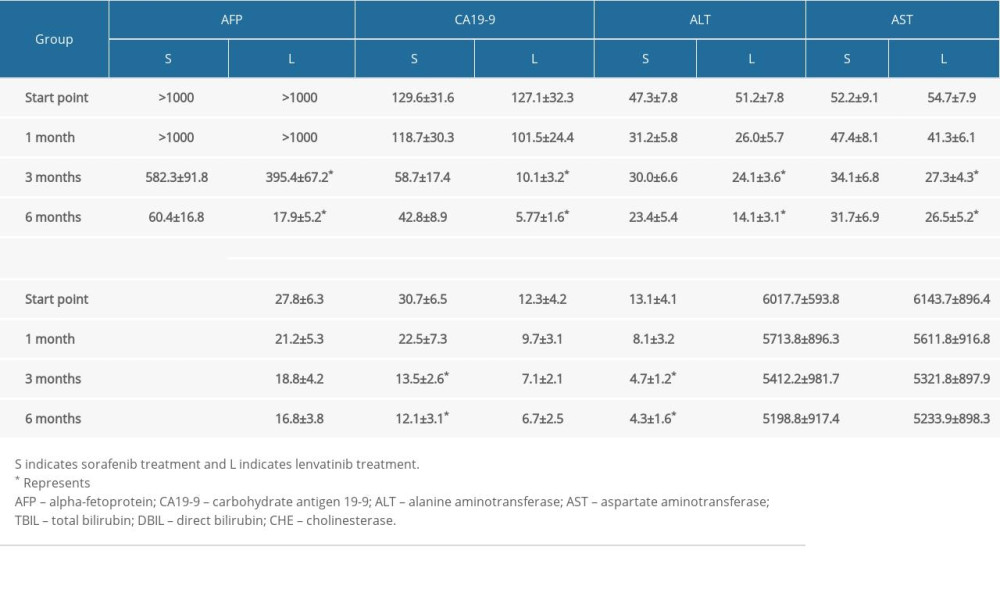 Table 1. Comparison of tumor markers and hepatobiliary function between 2 groups.
Table 1. Comparison of tumor markers and hepatobiliary function between 2 groups.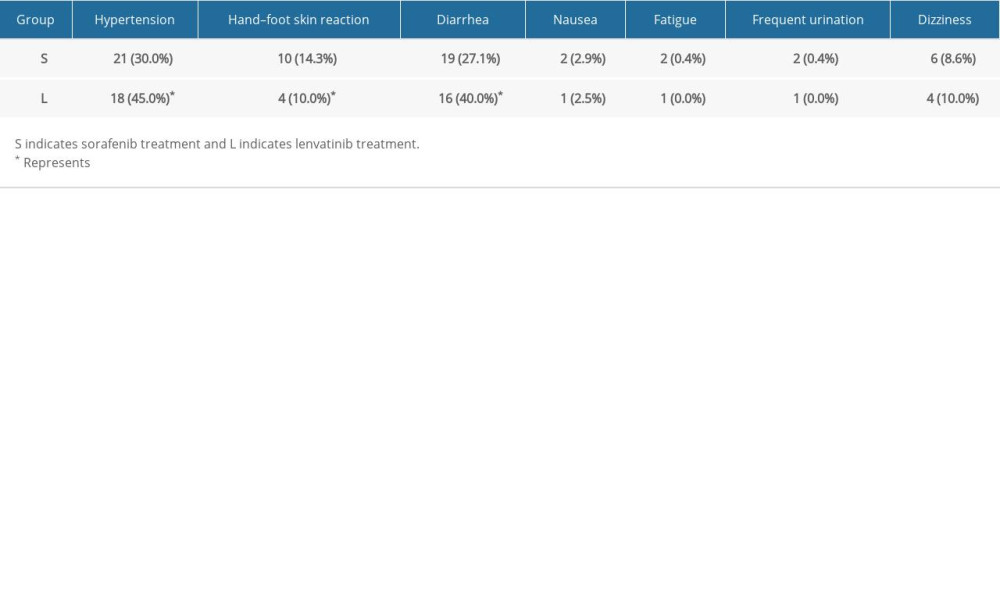 Table 2. Comparison of safety and adverse reactions of the 2 groups (case%).
Table 2. Comparison of safety and adverse reactions of the 2 groups (case%). Table 1. Comparison of tumor markers and hepatobiliary function between 2 groups.
Table 1. Comparison of tumor markers and hepatobiliary function between 2 groups. Table 2. Comparison of safety and adverse reactions of the 2 groups (case%).
Table 2. Comparison of safety and adverse reactions of the 2 groups (case%). In Press
05 Mar 2024 : Clinical Research
Muscular Function Recovery from General Anesthesia in 132 Patients Undergoing Surgery with Acceleromyograph...Med Sci Monit In Press; DOI: 10.12659/MSM.942780
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








